Revisiting guidance on population sampling for highly polymorphic STR loci
- PMID: 40779965
- PMCID: PMC12382342
- DOI: 10.1016/j.fsigen.2025.103336
Revisiting guidance on population sampling for highly polymorphic STR loci
Abstract
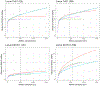
Population databases allow us to attach probabilities to DNA evidence by the estimation of genotype frequencies, which rely on accurate allele frequency estimates. As short tandem repeat (STR) marker sets for human identification have expanded to include more discriminating markers, and especially now that sequencing techniques allow us to distinguish between alleles based on variation in underlying base-pair structure, it is important to reevaluate existing guidance on population database sizes for the estimation of allele frequencies. In this paper, we revisit the topic of population sampling by focusing on the representation of alleles, i.e. whether alleles are observed or not, in a sample of individuals containing data for highly polymorphic autosomal STR loci. The effect of both length- and sequence-based STR data on population sample size implications are demonstrated, and differences between lesser and more polymorphic markers are discussed. The consequences of using a limited number of individuals are explored and the impact of increasing population sample sizes by combining different data sets is shown to help determine the point at which further sampling may no longer provide significant value. Finally, different approaches for accommodating previously unobserved alleles and their impact on DNA evidence evaluations are discussed.
Keywords: Allele frequency estimation; Forensic sequence data; Population sample size; Population studies.
Copyright © 2025 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
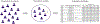





References
-
- Chakraborty R, Sample size requirements for addressing the population genetic issues of forensic use of DNA typing, Hum. Biol 64 (2) (1992) 141–159. - PubMed
-
- National Research Council. (1992). DNA Technology in Forensic Science. Washington, DC: National Academies Press. - PubMed
-
- Buckleton JS, Bright J, & Taylor D. (2016). Forensic DNA Evidence Interpretation (Second Edition). CRC Press.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

