ALDH1A3 promotes aggressive basal-like pancreatic cancer through an AP-1/RUNX2 enhancer network
- PMID: 40781158
- PMCID: PMC12477051
- DOI: 10.1038/s41388-025-03530-w
ALDH1A3 promotes aggressive basal-like pancreatic cancer through an AP-1/RUNX2 enhancer network
Abstract
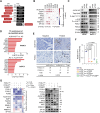
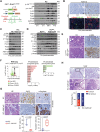
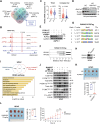
The basal-like transcriptional subtype of pancreatic ductal adenocarcinoma (PDAC) is linked to therapy resistance and poor prognosis. The cancer stem cell marker aldehyde dehydrogenase 1A3 (ALDH1A3) is a critical enzyme in acetaldehyde metabolism, but the interconnection to the basal-like subtype is poorly understood. Here, we identified ALDH1A3 as a key gene, which correlates with reduced survival and increased tumor growth. Functional studies revealed interaction of ALDH1A3 with genes like FAM3C, MCC, PMEPA1, and IRS2, forming a network driving PDAC progression. Chromatin profiling showed that ALDH1A3 affects acetylation of histone 3, mediating AP-1 activity, particularly via FOS family members, activating oncogenic pathways such as MAPK and TNF signaling. RUNX2 emerged as a therapeutic target within this network, as its knockdown disrupted MAPK signaling and reduced tumor growth. These findings emphasize the role of ALDH1A3 in linking nuclear metabolic-epigenetic programming in basal-like PDAC, highlighting it as a promising therapeutic target for novel treatment strategies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics: Clinical samples used in this study were approved by the Ethics Committee of the Technical University Munich (approval number: 80/17S) and the Ethics Committee of the Affiliated Drum Tower Hospital of Nanjing University (approval number: 2019-015-01). All mouse experiments and procedures were approved by the Institutional Animal Care and Use Committee of the Technical University of Munich (approval numbers: 55.2-1-54-2532-197-2016, ROB-55.2-2532.Vet_02-17-83), which address the ARRIVE guidelines. All procedures involving transgenic mice were performed in accordance with the Office of Laboratory Animal Welfare and German Federal Animal Protection Laws. BALB/c nu/nu athymic mice were approved by the Animal Care and Use Committee of the Nanjing Drum Tower Hospital (approval number: 20180102). All methods were performed in accordance with the relevant guidelines and regulations.
Figures



References
-
- Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17–48. 10.3322/caac.21763. - PubMed
-
- Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. 10.1038/nature16965. - PubMed
-
- Chan-Seng-Yue M, Kim JC, Wilson GW, Ng K, Figueroa EF, O’Kane GM, et al. Transcription phenotypes of pancreatic cancer are driven by genomic events during tumor evolution. Nat Genet. 2020;52:231–40. 10.1038/s41588-019-0566-9. - PubMed
MeSH terms
Substances
Grants and funding
- 70113828, 70115646/Deutsche Krebshilfe (German Cancer Aid)
- DFG KO 4369/4-1, KO 4369/6-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 2022.135.1/Wilhelm Sander-Stiftung (Wilhelm Sander Foundation)
- 82303254/National Natural Science Foundation of China (National Science Foundation of China)
- 8207265, 8207265, 81871947, 81602147/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

