Prediction of human pathogenic start loss variants based on self-supervised contrastive learning
- PMID: 40781627
- PMCID: PMC12333246
- DOI: 10.1186/s12915-025-02348-y
Prediction of human pathogenic start loss variants based on self-supervised contrastive learning
Abstract
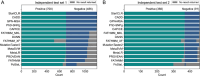
Background: Start loss variants are a class of genetic variants that affect the bases of the start codon, disrupting the normal translation initiation process and leading to protein deletions or the production of different proteins. Accurate assessment of the pathogenicity of these variants is crucial for deciphering disease mechanisms and integrating genomics into clinical practice. However, among the tens of thousands of start loss variants in the human genome, only about 1% have been classified as pathogenic or benign. Computational methods that rely solely on small amounts of labeled data often lack sufficient generalization capabilities, restricting their effectiveness in predicting the impact of start loss variants.
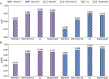
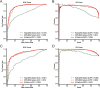
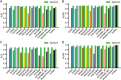
Results: Here, we introduce StartCLR, a novel prediction method specifically designed for identifying pathogenic start loss variants. StartCLR captures variant context information from different dimensions by integrating embedding features from diverse DNA language models. Moreover, it employs self-supervised pre-training combined with supervised fine-tuning, enabling the effective utilization of both a large amount of unlabeled data and a small amount of labeled data to enhance prediction accuracy. Our experimental results show that StartCLR exhibits strong generalization and superior prediction performance across different test sets. Notably, when trained exclusively on high-confidence labeled data, StartCLR retains or even improves the prediction accuracy despite the reduced amount of labeled data.
Conclusions: Collectively, these findings highlight the potential of integrating self-supervised contrastive learning with unlabeled data to mitigate the challenge posed by the scarcity of labeled start loss variants.
Keywords: Fine-tune; Pathogenicity prediction; Self-supervised contrastive learning; Start loss variant.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

