Exo-Tox: Identifying Exotoxins from secreted bacterial proteins
- PMID: 40781630
- PMCID: PMC12333140
- DOI: 10.1186/s13040-025-00469-2
Exo-Tox: Identifying Exotoxins from secreted bacterial proteins
Abstract
Background: Bacterial exotoxins are secreted proteins able to affect target cells, and associated with diseases. Their accurate identification can enhance drug discovery and ensure the safety of bacteria-based medical applications. However, current toxin predictors prioritize broad coverage by mixing toxins from multiple biological kingdoms and diverse control sets. This general approach has proven sub-optimal for identifying niche toxins, such as bacterial exotoxins. Recent Protein Language Models offer an opportunity to improve toxin prediction by capturing global sequence context and biochemical properties from protein sequences.
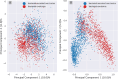
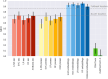
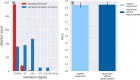
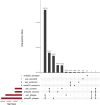
Results: We introduce Exo-Tox, a specialized predictor trained exclusively on curated datasets of bacterial exotoxins and secreted non-toxic bacterial proteins, represented as embeddings by Protein Language Models. Compared to Basic Local Alignment Search Tool (BLAST)-based methods and generalized toxin predictors, Exo-Tox outperforms across multiple metrics, achieving a Matthews correlation coefficient > 0.9. Notably, Exo-Tox's performance remains robust regardless of protein length or the presence of signal peptides. We analyze its limited transferability to bacteriophage proteins and non-secreted proteins.
Conclusion: Exo-Tox reliably identifies bacterial exotoxins, filling a niche overlooked by generalized predictors. Our findings highlight the importance of domain-specific training data and emphasize that specialized predictors are necessary for accurate classification. We provide open access to the model, training data, and usage guidelines via the LMU Munich Open Data repository.
Keywords: Bacterial; Embeddings; Exotoxins; Predictor; Proteins; Toxins.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures
Similar articles
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Sexual Harassment and Prevention Training.2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 36508513 Free Books & Documents.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Leveraging a foundation model zoo for cell similarity search in oncological microscopy across devices.Front Oncol. 2025 Jun 18;15:1480384. doi: 10.3389/fonc.2025.1480384. eCollection 2025. Front Oncol. 2025. PMID: 40606969 Free PMC article.
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

