Mesenchymal stromal cells conditioned by peripheral blood mononuclear cells exert enhanced immunomodulation capacities and alleviate a model of Myasthenia Gravis
- PMID: 40781689
- PMCID: PMC12333171
- DOI: 10.1186/s13287-025-04534-9
Mesenchymal stromal cells conditioned by peripheral blood mononuclear cells exert enhanced immunomodulation capacities and alleviate a model of Myasthenia Gravis
Abstract
Background: Mesenchymal Stromal Cells (MSC) possess innate immunomodulatory properties, which can be significantly enhanced through co-culture with peripheral blood mononuclear cells (PBMC), making them attractive tools for the treatment of autoimmune and inflammatory diseases.
Methods: Leveraging a multi-omics approach encompassing RNA sequencing, flow and mass cytometry, secretome analysis, completed by functional evaluations, we investigated the mechanisms underpinning PBMC conditioning of MSC in vitro and their benefits in an animal model of Myasthenia gravis. MSC derived from human adipose tissue were left untreated in resting state (rMSC), conditioned by PBMC (cMSC), or activated by the pro-inflammatory molecule interferon (IFN)-γ (γMSC), then compared for their gene expression profiles, phenotypes and functional capacities.
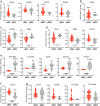
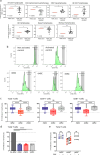
Results: RNA sequencing identified 244 differentially expressed genes in cMSC compared to rMSC, highlighting key immune mediators such as CCL2, CCL11, DPP4, ICAM1, IL6, PDCD1LG2, TNFRSF11B, TNIP1, TNIP3 and ZC3H12A and pinpointing genes involved in matrix remodeling, paracrine and autocrine communications. Comparatively, 2089 genes were differentially expressed between rMSC and γMSC, highlighting host defense, anti-viral response, NFκB signaling pathways modulated by IFN-γ. Flow and mass cytometry analyses revealed upregulation of the surface markers CD26, CD54, and CD273 and intracellular molecules IDO1 and PTGS2 in cMSC. In contrast, IFN-γ activation predominantly increased HLA-related markers while also enhancing the homogeneity of the populations. Together, these results underlined the treatment dependence of transcriptomic and phenotypic signatures. Secretome profiling identified 6 categories of modulated proteins, out of which 22 molecules potentially involved in PBMC conditioning and 40 implicated in cMSC-mediated immunomodulation. Functionally, cMSC induced modulation in PBMC subsets, raising the proportions of lymphocyte populations (CD4 Treg, CD8, B memory), underlining the multimodal effect of conditioning. Also, both a direct cell-cell contact and cMSC supernatants significantly suppressed activated T-cell proliferation in vitro. To confirm immunomodulation efficacy in vivo, cMSC were administrated to our humanized mouse model of Myasthenia Gravis and the treatment significantly halved disease severity from 2 weeks post-injection.
Conclusions: This integrative study establishes distinct conditioning signatures, suggests molecular mechanisms, and underscores the therapeutic potential of cMSC, offering a robust framework for advancing cell-based therapies in autoimmune diseases.
Keywords: Conditioning; Humanized mouse model; Immunomodulation; Inhibition of proliferation; Mass cytometry; Mesenchymal stromal cells; Myasthenia gravis; RNASeq; Secretome; γ-Interferon.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Human adipose-derived MSC were obtained in the context of two successive clinical trials and used as res nullius. (1) The title of the aproved projects were ADIPOA (A Phase I, Prospective, bi-centric, single-arm, open-label, dose-escalating clinical trial to evaluate the safety of a single injection of autologous adipose derived mesenchymal stromal cells in the treatment of severe osteoarthritis of the nee joint) and ADIPOA-2 (A study evaluating the efficacy of a single injection of autologous adipose derived mesenchymal stromal cells in patients with knee osteoarthritis). (2) They were authorized by the Personal Protection Committee of Montpellier, France, UF 606-120203. (3) The registered numbers are NCT01585857 and NCT02838069. (4) The projects were approved on 12/2011 and 09/2016. According to the French laws and regulations, the patients provided written informed consent for use of their cells and were anonymized. AChR+MG thymic fragments were provided by medical collaborators with the authorization of a French Ethical Committee. (1) The title of the approved project was MYA-PATH (Constitution of a collection of biological resources for the study of pathologies that could lead to autoimmune myasthenia). (2) It was authorized by the Personal Protection Committee of Tours, Central West Region 1, France, IRB n° IORG0008143 OMB: 0990 − 0279, (3) under agreement number 2019-A01482-55, (4) in date of 11/20/2019. The patients were undergoing thymectomy and gave written informed consent for the use of res nullius samples obtained during surgeries and anonymized according to the French laws and regulations. Whole blood bags were procured by the French Blood Establishment (EFS Ile de France) to prepare PBMC according to the convention CCPSL UNT– N° 18/EFS/033 authorized on 16/08/2018 between our Institutions. According to French laws and regulations (article R.1222-40/2 of the Public Health Code), the EFS collect and distribute blood elements prepared from donors, whose gift is free and anonymous, and who gave written informed consent for their use. No further IRB was required. Animal experimentations have been designed to respect the 3R principles according to the Directive 2010/63/EU of the European Union and received the APAFIS authorizations number 02622.2 (Evaluation of cellular and pharmacological therapeutic strategies in the experimental mouse model of Myasthenia gravis, 20/11/2015) and number 30172 (Set up of cellular therapy strategies in two experimental mouse models of Myasthenia gravis, 21/05/2021), from the French Ministry of Higher Education and Research under agreement of the local Ethical Committee (CEEA 005, Charles Darwin). Experimentations were performed under supervision of personals authorized by French Authorities in the animal facility of the Sorbonne University (EU 0144, Agreement C-75-13-20). Consent for publication: Not applicable. Competing interests: The authors declare that they have no competing financial or personal interests that could have influenced the work reported in this article.
Figures






Similar articles
-
Extracellular vesicles derived from clonal mesenchymal stromal cells preconditioned by indirect hypoxia modulate immune responses in diabetic mice more effectively than directly preconditioned vesicles.Stem Cell Res Ther. 2025 Aug 26;16(1):458. doi: 10.1186/s13287-025-04568-z. Stem Cell Res Ther. 2025. PMID: 40859321 Free PMC article.
-
In-vitro immunomodulatory efficacy of extracellular vesicles derived from TGF-β1/IFN-γ dual licensed human bone marrow mesenchymal stromal cells.Stem Cell Res Ther. 2025 Jul 9;16(1):357. doi: 10.1186/s13287-025-04476-2. Stem Cell Res Ther. 2025. PMID: 40629458 Free PMC article.
-
Mesenchymal stem cells and their secretome modulate stress and enhance lipogenesis in bovine mammary epithelial cells.Stem Cell Res Ther. 2025 Jun 23;16(1):320. doi: 10.1186/s13287-025-04442-y. Stem Cell Res Ther. 2025. PMID: 40551173 Free PMC article.
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
-
- Andreeva E, Bobyleva P, Gornostaeva A, Buravkova L. Interaction of multipotent mesenchymal stromal and immune cells: bidirectional effects. Cytotherapy. 2017;19(10):1152–66. - PubMed
-
- Ben-Ami E, Berrih-Aknin S, Miller A. Mesenchymal stem cells as an Immunomodulatory therapeutic strategy for autoimmune diseases. Autoimmun Rev. 2011;10(7):410–5. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

