CDH3 promotes the progression of lung adenocarcinoma through driving epithelial-mesenchymal transition progress
- PMID: 40781712
- PMCID: PMC12333065
- DOI: 10.1186/s12967-025-06750-6
CDH3 promotes the progression of lung adenocarcinoma through driving epithelial-mesenchymal transition progress
Abstract
Background: Cadherin-3 (CDH3) participates in multiple oncogenic processes, but its biological role and mechanisms in lung adenocarcinoma (LUAD) remain inadequately understood.
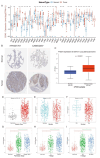
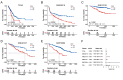
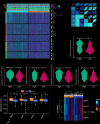
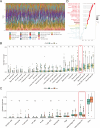
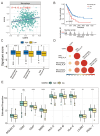
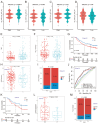
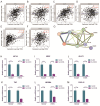
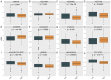
Methods: We analyzed CDH3 expression in LUAD versus adjacent normal tissues and assessed its association with patient outcomes. Bioinformatics, immune scoring, and single-cell analysis were used to explore CDH3's links to epithelial-mesenchymal transition (EMT), inflammatory responses, TNF pathway activation, glycolysis, hypoxia, and tumor-associated macrophage infiltration. Independent immunotherapy datasets (IMvigor210, GSE91061) were evaluated for CDH3's relationship to treatment response. In vitro CDH3 knockdown studies assessed effects on glycolysis, EMT, proliferation, and migration in LUAD cells, and in vivo tumor growth was measured post-knockdown.
Results: CDH3 expression was significantly elevated in LUAD tissues and correlated with poorer patient survival. High CDH3 expression associated with EMT, inflammatory responses (including TNF pathway), glycolysis, and hypoxia. It also promoted tumor-associated macrophage infiltration and impaired anti-tumor immunity. Patients with low CDH3 expression showed improved immunotherapy response and prognosis. CDH3 knockdown suppressed LUAD cells glycolysis process, EMT, proliferation, and migration in vitro, and significantly repressed tumor growth in vivo.
Conclusions: CDH3 drives LUAD progression by promoting EMT, tumor growth, metastasis, and immunosuppression. It represents a promising diagnostic/prognostic biomarker and a novel therapeutic target for LUAD immunotherapy.
Keywords: CDH3; EMT; Immunotherapy; Lung adenocarcinoma; Tumor associated macrophages.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare that the study was implemented without any financial or commercial correlations that could be established as underlying conflict of interest.
Figures



Similar articles
-
Caveolin-1 inhibits the proliferation and invasion of lung adenocarcinoma via EGFR degradation.Sci Rep. 2025 Jul 1;15(1):21654. doi: 10.1038/s41598-025-05259-8. Sci Rep. 2025. PMID: 40594106 Free PMC article.
-
Unraveling the role of GPCR signaling in metabolic reprogramming and immune microenvironment of lung adenocarcinoma: a multi-omics study with experimental validation.Front Immunol. 2025 Jun 6;16:1606125. doi: 10.3389/fimmu.2025.1606125. eCollection 2025. Front Immunol. 2025. PMID: 40547013 Free PMC article.
-
SMC2 knockdown inhibits malignant progression of lung adenocarcinoma by upregulating BTG2 expression.Cell Signal. 2024 Aug;120:111216. doi: 10.1016/j.cellsig.2024.111216. Epub 2024 May 8. Cell Signal. 2024. PMID: 38729325
-
UBE2N as a novel prognostic and therapeutic biomarker of lung adenocarcinoma.Front Immunol. 2025 Aug 11;16:1636503. doi: 10.3389/fimmu.2025.1636503. eCollection 2025. Front Immunol. 2025. PMID: 40861472 Free PMC article.
-
CENPA-driven transcriptional activation of ECT2 enhances EGFR inhibitor resistance in lung adenocarcinoma.Biochem Biophys Res Commun. 2025 Sep 1;777:152287. doi: 10.1016/j.bbrc.2025.152287. Epub 2025 Jul 3. Biochem Biophys Res Commun. 2025. PMID: 40618443
References
MeSH terms
Substances
Grants and funding
- 82122053/National Natural Science Foundation of China
- 021-1-I2M-067/Chinese Academy of Medical Sciences Initiative for Innovative Medicine
- 2021B0101420005/Key-Area Research and Development Program of Guangdong Province
- ZDSYS20220606101604009/Shenzhen Science and Technology Innovation Program
- SZSM202211011/Sanming Project of Medicine in Shenzen Municipality
LinkOut - more resources
Full Text Sources
Medical

