Targeting NOTCH1-KEAP1 axis retards chronic liver injury and liver cancer progression via regulating stabilization of NRF2
- PMID: 40781716
- PMCID: PMC12335071
- DOI: 10.1186/s13046-025-03488-3
Targeting NOTCH1-KEAP1 axis retards chronic liver injury and liver cancer progression via regulating stabilization of NRF2
Abstract
Background: Chronic liver injury is a key factor in diseases like hepatocellular carcinoma (HCC), steatohepatitis (NASH), and viral hepatitis type B and C (HBV, HCV). Understanding its molecular mechanisms is crucial for effective treatment. The NOTCH1 signaling pathway, though not fully understood, is implicated in liver injury and may be a potential therapeutic target.
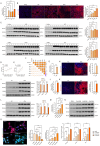
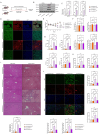
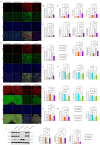
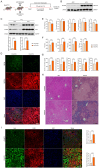
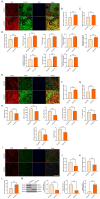
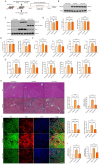
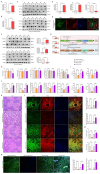
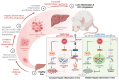
Methods: Clinical HCC, HBV, HCV and NASH samples and additional in vitro and in vivo performances were subjected to confirm the role of NOTCH1 and its downstream targets via a series of biochemical assays, molecular analysis approaches and targeted signaling pathway assay, etc. RESULTS: The present study first verified the abnormal elevation of NOTCH1 in hepatocytes from patients with steatohepatitis, HCC, HBV, HCV, and mouse models. Crucially, we discovered that hepatocyte-specific NOTCH1 knockout reduces hepatocellular damage in chronic liver inflammation and HCC mouse models, whereas adeno-associated virus serotype 8 (AAV8)-mediated NOTCH1 overexpression in hepatocytes exacerbates liver injury-related phenotype on-setting. Mechanistically, we showed that NOTCH1 has a new role in controlling ferroptosis and oxidative damage in hepatocytes. It interacts with Kelch-like ECH-associated protein 1 (KEAP1) and is directly recruited through its intracellular domain (NICD1). Additionally, the KEAP1 recruited by NOTCH1 impeded the binding stability of KEAP1-NFE2 like BZIP transcription factor 2 (Nrf2), promote the separation of KEAP1 and Nrf2, thereby reducing the stability of Nrf2 and hindering the ubiquitination-related proteasome degradation of Nrf2. Crucially, we also discovered that NOTCH1's ANK domain is essential for NICD1-KEAP1 contacts and signaling activation. The inability of NOTCH1 with ANK domain mutants (ΔANK) to connect with KEAP1 and increase its expression emphasizes the importance of the ANK domain in KEAP1-NRF2 signaling. By reversing the downregulation of KEAP1 and the overexpression of NRF2, ANK function is linked to ferroptosis and ROS buildup. ANK domain targeting may slow the course of HCC and liver damage.
Conclusions: Targeting the NOTCH1-KEAP1-NRF2 axis as a possible chronic hepatic injury therapy is supported by these findings, which identify NOTCH1-KEAP1 as an NRF2 suppressor that accelerates the progression of liver injury.
Keywords: Chronic liver inflammation; Ferroptosis; Hepatocellular carcinoma (HCC); NOTCH1; Nrf2.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The use of clinical samples was approved by the ethics committee of Shandong First Medical University & Shandong Academy of Medical Sciences and informed consent were obtained from all patients (2024LC0001). All animal experimental protocols were performed following the Ethical Animal Care and Use Committee of Shandong University (2022JJCA08). Consent for publication: All authors consent to the publication of this article. Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
CYP4F11, an NRF2 Target Gene, Promotes Hepatocellular Carcinoma Cell Growth.Mol Carcinog. 2025 Jul;64(7):1264-1274. doi: 10.1002/mc.23925. Epub 2025 May 6. Mol Carcinog. 2025. PMID: 40329467
-
Competitive Binding of Pseudomonas aeruginosa to Kelch-Like ECH-Associated Protein 1 Inhibits Nuclear Factor Erythroid 2-Related Factor 2 Ubiquitination and Suppresses Ferroptosis in Macrophages.Microbiol Immunol. 2025 Aug;69(8):418-428. doi: 10.1111/1348-0421.13228. Epub 2025 Jun 4. Microbiol Immunol. 2025. PMID: 40468416
-
Fbxw7 regulates hepatocellular carcinoma migration and invasion via Notch1 signaling pathway.Int J Oncol. 2015 Jul;47(1):231-43. doi: 10.3892/ijo.2015.2981. Epub 2015 May 4. Int J Oncol. 2015. Retraction in: Int J Oncol. 2025 Jul;67(1):57. doi: 10.3892/ijo.2025.5763. PMID: 25955618 Retracted.
-
Nonalcoholic fatty liver disease and hepatocellular carcinoma.Metabolism. 2016 Aug;65(8):1151-60. doi: 10.1016/j.metabol.2016.01.010. Epub 2016 Jan 23. Metabolism. 2016. PMID: 26907206
-
Metal profiling in coronary ischemia-reperfusion injury: Implications for KEAP1/NRF2 regulated redox signaling.Free Radic Biol Med. 2024 Jan;210:158-171. doi: 10.1016/j.freeradbiomed.2023.11.013. Epub 2023 Nov 19. Free Radic Biol Med. 2024. PMID: 37989446 Review.
References
-
- Devarbhavi H, Asrani SK, Arab JP, Nartey YA, Pose E, Kamath PS. Global burden of liver disease: 2023 update. J Hepatol. 2023;79(2):516–37. - PubMed
-
- Hammerich L, Tacke F. Hepatic inflammatory responses in liver fibrosis. Nat Reviews Gastroenterol Hepatol. 2023;20(10):633–46. - PubMed
-
- Xu M, Zhao J, Zhu L, Ge C, Sun Y, Wang R, Shi S. Targeting PYK2 with heterobifunctional T6BP helps mitigate MASLD and MASH-HCC progression. J Hepatol. 2025;82(2):277–300. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

