Microbial diversity and functional potential of the Halobates melleus (Heteroptera: Gerridae) microbiome from the Red Sea coastline
- PMID: 40781723
- PMCID: PMC12335105
- DOI: 10.1186/s40793-025-00761-y
Microbial diversity and functional potential of the Halobates melleus (Heteroptera: Gerridae) microbiome from the Red Sea coastline
Abstract
Background: Halobates, commonly known as sea skaters, are predatory Hemipterans uniquely adapted to tropical marine environments. Their ability to thrive in oligotrophic and environmentally extreme habitats, such as the open ocean surface and marine coastal areas, suggests the evolution of specialised adaptations, possibly including symbiotic associations with microorganisms that can support nutrition, niche adaptation, and stress resilience. To explore this hypothesis, we analysed the bacterial communities associated with Halobates melleus, a species inhabiting the Red Sea coastal mangroves in Saudi Arabia.
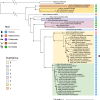
Results: Amplicon sequencing of the 16S rRNA gene and metagenomic analyses of composite body and gut samples from adult H. melleus revealed a population-level bacterial community dominated by Wolbachia and Spiroplasma, consistent with patterns observed in several terrestrial predatory insects. Members of Providencia and Swaminathania were also detected, along with other minor taxa that may represent transient environmental commensals. The identified bacteria encoded genes for the biosynthesis of essential vitamins and prosthetic groups, such as riboflavin and heme-compounds typically not synthesised de novo by insects-as well as amino acids, likely contributing to the host's nutritional requirements. Notably, the Wolbachia metagenome-assembled genome from H. melleus clustered within the supergroup B, showing high genetic similarity to strains from phylogenetically distant Dipteran and Lepidopteran hosts that nonetheless inhabit common ecological niches, i.e., mangrove and tropical environments. This extends the known ecological breadth of Wolbachia symbioses to marine insects, underscoring their evolutionary and environmental versatility.
Conclusion: Our findings highlight the potential nutritional and metabolic roles of the Halobates-associated bacterial microbiome, particularly members of the Wolbachia genus. This emphasises the importance of microbial symbionts in the ecological success and adaptation of marine insects, offering a perspective complementary to previously studied terrestrial insect microbiomes.
Keywords: Halobates; Wolbachia; Extreme environment; Marine insect; Metabolic potential; Metagenome; Microbiome; Symbiont.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interest: The authors declare no competing interests.
Figures




Similar articles
-
Ecological differences among hydrothermal vent symbioses may drive contrasting patterns of symbiont population differentiation.mSystems. 2023 Aug 31;8(4):e0028423. doi: 10.1128/msystems.00284-23. Epub 2023 Jul 26. mSystems. 2023. PMID: 37493648 Free PMC article.
-
Extremely distinct microbial communities in closely related leafhopper subfamilies: Typhlocybinae and Eurymelinae (Cicadellidae, Hemiptera).mSystems. 2025 Jul 22;10(7):e0060325. doi: 10.1128/msystems.00603-25. Epub 2025 Jun 26. mSystems. 2025. PMID: 40569073 Free PMC article.
-
Phylogenetic and functional diversity among Drosophila-associated metagenome-assembled genomes.mSystems. 2025 Jul 22;10(7):e0002725. doi: 10.1128/msystems.00027-25. Epub 2025 Jul 2. mSystems. 2025. PMID: 40600712 Free PMC article.
-
Factors that influence parents' and informal caregivers' views and practices regarding routine childhood vaccination: a qualitative evidence synthesis.Cochrane Database Syst Rev. 2021 Oct 27;10(10):CD013265. doi: 10.1002/14651858.CD013265.pub2. Cochrane Database Syst Rev. 2021. PMID: 34706066 Free PMC article.
-
Measures implemented in the school setting to contain the COVID-19 pandemic.Cochrane Database Syst Rev. 2022 Jan 17;1(1):CD015029. doi: 10.1002/14651858.CD015029. Cochrane Database Syst Rev. 2022. Update in: Cochrane Database Syst Rev. 2024 May 2;5:CD015029. doi: 10.1002/14651858.CD015029.pub2. PMID: 35037252 Free PMC article. Updated.
References
-
- Andersen NM, Cheng L. The marine insect Halobates (Heteroptera: Gerridae): biology, adaptations, distribution, and phylogeny. Oceanogr Mar Biol Annu Rev. 2004;42:119–80.
-
- Cheng L, Baars M, Smith A. Life on the high seas- the bug Darwin never saw: Introduction and historical background. Antenna. 2011;35:36–42.
-
- Cheng L. Biology of Halobates (Heteroptera: Gerridae). Annu Rev Entomol. 1985;30:111–35.
Grants and funding
LinkOut - more resources
Full Text Sources
