Combined MEK and PARP inhibition enhances radiation response in rectal cancer
- PMID: 40782795
- PMCID: PMC12432356
- DOI: 10.1016/j.xcrm.2025.102284
Combined MEK and PARP inhibition enhances radiation response in rectal cancer
Abstract
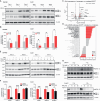
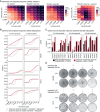
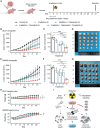
Rectal cancer is frequently diagnosed at a locally advanced stage and treated by neoadjuvant chemoradiation. Current efforts to improve treatment outcome are focused on intensifying neoadjuvant chemotherapy, which is associated with higher levels of toxicity. To discover alternative strategies, we establish patient-derived rectal cancer organoids that reflect clinical radiosensitivity and use these organoids to screen 1,596 drug-radiation combinations. We find that inhibitors of rat sarcoma virus/mitogen-activated protein kinase (RAS-MAPK) signaling, especially mitogen-activated protein kinase kinase (MEK) inhibitors, strongly enhance radiation response. Mechanistically, MEK inhibitors suppress radiation-induced activation of RAS-MAPK signaling and selectively downregulate RAD51, a component of the homologous recombination DNA repair pathway. Through testing drug-drug-radiation combinations in organoids and cell lines, we identify that a combined poly ADP-ribose polymerase (PARP) and MEK inhibition can further enhance radiosensitivity of colorectal cancers, which we confirm in mouse xenograft models. Our data support clinical testing of MEK and PARP combination therapy with radiation in locally advanced rectal cancers as an alternative to chemoradiation.
Keywords: DNA repair; MEK inhibitor; PARP inhibitor; RAD51; RAS signaling; drug combination screen; organoids; radiation therapy; radiosensitizer; rectal cancer.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Sauer R., Liersch T., Merkel S., Fietkau R., Hohenberger W., Hess C., Becker H., Raab H.-R., Villanueva M.-T., Witzigmann H., et al. Preoperative Versus Postoperative Chemoradiotherapy for Locally Advanced Rectal Cancer: Results of the German CAO/ARO/AIO-94 Randomized Phase III Trial After a Median Follow-Up of 11 Years. J. Clin. Oncol. 2012;30:1926–1933. doi: 10.1200/jco.2011.40.1836. - DOI - PubMed
-
- Bahadoer R.R., Dijkstra E.A., van Etten B., Marijnen C.A.M., Putter H., Kranenbarg E.M.-K., Roodvoets A.G.H., Nagtegaal I.D., Beets-Tan R.G.H., Blomqvist L.K., et al. Short-course radiotherapy followed by chemotherapy before total mesorectal excision (TME) versus preoperative chemoradiotherapy, TME, and optional adjuvant chemotherapy in locally advanced rectal cancer (RAPIDO): a randomised, open-label, phase 3 trial. Lancet Oncol. 2021;22:29–42. doi: 10.1016/s1470-2045(20)30555-6. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

