Mitochondrial damage triggers the concerted degradation of negative regulators of neuronal autophagy
- PMID: 40783388
- PMCID: PMC12335601
- DOI: 10.1038/s41467-025-62379-5
Mitochondrial damage triggers the concerted degradation of negative regulators of neuronal autophagy
Abstract
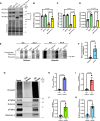
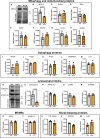
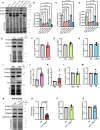
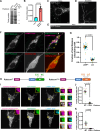
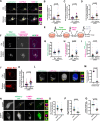
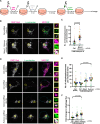
Mutations that disrupt the clearance of damaged mitochondria via mitophagy are causative for neurological disorders including Parkinson's. Here, we identify a Mitophagic Stress Response (MitoSR) activated by mitochondrial damage in neurons and operating in parallel to canonical Pink1/Parkin-dependent mitophagy. Increasing levels of mitochondrial stress trigger a graded response that induces the concerted degradation of negative regulators of autophagy including Myotubularin-related phosphatase (MTMR)5, MTMR2 and Rubicon via the ubiquitin-proteasome pathway and selective proteolysis. MTMR5/MTMR2 inhibit autophagosome biogenesis; consistent with this, mitochondrial engulfment by autophagosomes is enhanced upon MTMR2 depletion. Rubicon inhibits lysosomal function, blocking later steps of neuronal autophagy; Rubicon depletion relieves this inhibition. Targeted depletion of both MTMR2 and Rubicon is sufficient to enhance mitophagy, promoting autophagosome biogenesis and facilitating mitophagosome-lysosome fusion. Together, these findings suggest that therapeutic activation of MitoSR to induce the selective degradation of negative regulators of autophagy may enhance mitochondrial quality control in stressed neurons.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Vakifahmetoglu-Norberg, H., Ouchida, A. T. & Norberg, E. The role of mitochondria in metabolism and cell death. Biochem. Biophys. Res. Commun.482, 426–431 (2017). - PubMed
-
- Rangaraju, V., Lauterbach, M. & Schuman, E. M. Spatially stable mitochondrial compartments fuel local translation during plasticity. Cell176, 73–84.e15 (2019). - PubMed
-
- Devine, M. J. & Kittler, J. T. Mitochondria at the neuronal presynapse in health and disease. Nat. Rev. Neurosci.19, 63–80 (2018). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

