Opposing regulation of the K63-linked polyubiquitination of RIPK3 by SMURF1 and USP5 in necroptosis
- PMID: 40783390
- PMCID: PMC12335473
- DOI: 10.1038/s41467-025-62723-9
Opposing regulation of the K63-linked polyubiquitination of RIPK3 by SMURF1 and USP5 in necroptosis
Abstract
Receptor-interacting protein kinase 3 (RIPK3), a key regulator of necroptosis, is modulated by ubiquitination through various E3 ligases and deubiquitinases. However, the effects of different polyubiquitination processes on RIPK3 and necroptosis remain unclear. Using a proteomic approach, we identify SMAD Ubiquitination Regulatory Factor 1 (SMURF1) and Ubiquitin-specific peptidase 5 (USP5) as crucial regulators of RIPK3 within the necrosome during necroptosis. SMURF1 facilitates K63 polyubiquitination of RIPK3 at lysine 55 and 363, inhibiting necrosome formation and necroptosis. SMURF1 depletion accelerates necroptosis, while the reintroduction of functional SMURF1 reverses this. Conversely, USP5 acts as a deubiquitinase, removing K63 ubiquitin chains and promoting necroptosis. Reducing SMURF1, using a RIPK3 mutant defective in SMURF1-mediated ubiquitination, or overexpressing USP5 enhances necroptosis in leukaemia cells, leading to reduced tumour growth in xenograft models treated with birinapant and emricasan. These findings highlight the opposing regulation of K63-linked polyubiquitination of RIPK3 by SMURF1 and USP5 in necroptosis.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
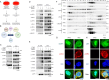



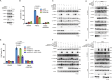


References
-
- Vandenabeele, P., Galluzzi, L., Vanden Berghe, T. & Kroemer, G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat. Rev. Mol. cell Biol.11, 700–714 (2010). - PubMed
-
- Newton, K. et al. Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis. Science343, 1357–1360 (2014). - PubMed
-
- Kaczmarek, A., Vandenabeele, P. & Krysko, D. V. Necroptosis: the release of damage-associated molecular patterns and its physiological relevance. Immunity38, 209–223 (2013). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

