Prx1+ progenitors give rise to new articular cartilage when conditions are permissive for endogenous regeneration
- PMID: 40783399
- PMCID: PMC12335530
- DOI: 10.1038/s41536-025-00425-y
Prx1+ progenitors give rise to new articular cartilage when conditions are permissive for endogenous regeneration
Abstract
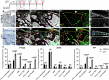
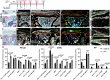
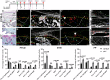
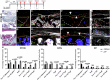
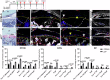
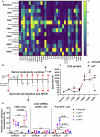
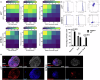
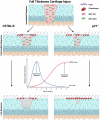
It is widely acknowledged that articular cartilage lacks the ability to regenerate. However, if such regeneration were possible, which cell type would generate new tissue? The p21-/- mouse provides an excellent platform to explore this question, hence, we conducted lineage tracing on Paired related homeobox 1 (Prrx1/Prx1) cells post-injury to determine whether endogenous Prx1+ cells contribute to regenerated tissues post-injury. p21-/- mice displayed enhanced endogenous cartilage regeneration, accompanied by notable differences in the number and kinetics of Prx1+ cells within and around the injury site. In p21-/- mice, Prx1+ cells underwent chondrogenesis, ultimately contributing to the regenerated articular cartilage layer. These findings underscore the impact of tissue-resident cells on cartilage regeneration, albeit under abnormal conditions. If the conditions within the joint could be manipulated to favor such a regenerative environment, these endogenous cell types might be recruited to facilitate the formation of a new articular cartilage surface post-injury.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
NFATc1 marks articular cartilage progenitors and negatively determines articular chondrocyte differentiation.Elife. 2023 Feb 15;12:e81569. doi: 10.7554/eLife.81569. Elife. 2023. PMID: 36790146 Free PMC article.
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Sexual Harassment and Prevention Training.2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 36508513 Free Books & Documents.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
-
Trocar types in laparoscopy.Cochrane Database Syst Rev. 2015 Dec 16;2015(12):CD009814. doi: 10.1002/14651858.CD009814.pub2. Cochrane Database Syst Rev. 2015. PMID: 26676093 Free PMC article.
References
-
- Hunter, W. VI. Of the structure and diseases of articulating cartilages. Philos. Trans. R. Soc. Lond.42, 514–521 (1743).
-
- Charalambous, C. P. The response of articular cartilage to mechanical injury. Class. Pap. Orthop.64, 381–383 (2014).
Grants and funding
- RGPIN-2014-04586/Gouvernement du Canada | Natural Sciences and Engineering Research Council of Canada (Conseil de Recherches en Sciences Naturelles et en Génie du Canada)
- Create Fellowship/Gouvernement du Canada | Natural Sciences and Engineering Research Council of Canada (Conseil de Recherches en Sciences Naturelles et en Génie du Canada)
- JELF/Canada Foundation for Innovation (Fondation canadienne pour l'innovation)
- 201309MOP-311542-DIB-CBBA-170939/Gouvernement du Canada | Canadian Institutes of Health Research (Instituts de Recherche en Santé du Canada)
- Grace Glaum Professorship/Calgary Foundation (CF)
LinkOut - more resources
Full Text Sources
Research Materials

