A multivalent mRNA vaccine elicits robust immune responses and confers protection in a murine model of monkeypox virus infection
- PMID: 40783493
- PMCID: PMC12335498
- DOI: 10.1038/s41467-025-61699-w
A multivalent mRNA vaccine elicits robust immune responses and confers protection in a murine model of monkeypox virus infection
Abstract
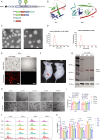
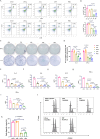
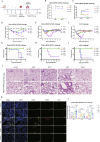
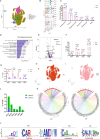
Monkeypox virus (MPXV) has re-emerged globally since May 2022, posing a significant public health threat. To address this, we develop two multivalent mRNA vaccine candidates-AAL, encoding three MPXV antigens, and AALI, which combines AAL with an immune-enhancing IFN-α protein. Both vaccines are delivered via mannose-modified lipid nanoparticles to target dendritic cells. Here we show that these vaccines elicit strong antibody responses against vaccinia virus and multiple MPXV clades, induce robust memory B-cell and T-cell responses, and promote dendritic cell maturation. In mouse challenge models, both vaccines provide protection against clade IIb MPXV and vaccinia virus, significantly reducing viral loads and preventing lung damage. Immune profiling reveals enhanced B- and T-cell receptor diversity and distinct CDR3 motifs post-vaccination. These findings demonstrate the potential of using mRNA-based multivalent vaccines as an effective strategy for preventing mpox and related Orthopoxvirus infections.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors S.C., S.Y.L., and L.W. have filed a Chinese patent application (number: 202410069336.7) for the MPXV mRNA vaccines and their construction method and application. The other authors do not claim competing interests.
Figures



Similar articles
-
Correlation between microneutralization test and a multiplexed immunoassay for evaluation of monkeypox and vaccinia virus antibodies before and after smallpox vaccination.Front Immunol. 2025 Jun 23;16:1585284. doi: 10.3389/fimmu.2025.1585284. eCollection 2025. Front Immunol. 2025. PMID: 40625755 Free PMC article.
-
Multiple gene-deletion vaccinia virus Tiantan strain against mpox.Virol J. 2025 Jan 25;22(1):17. doi: 10.1186/s12985-025-02629-6. Virol J. 2025. PMID: 39863910 Free PMC article.
-
A bivalent Mpox nanoparticle vaccine induces robust immune response and provides long-lasting protection against vaccinia virus challenge.Emerg Microbes Infect. 2025 Dec;14(1):2535485. doi: 10.1080/22221751.2025.2535485. Epub 2025 Jul 28. Emerg Microbes Infect. 2025. PMID: 40720261 Free PMC article.
-
Multi-epitope vaccines: charting a new frontier in monkeypox prevention and control.Hum Cell. 2025 Jul 9;38(5):126. doi: 10.1007/s13577-025-01255-2. Hum Cell. 2025. PMID: 40632349 Review.
-
Exploring monkeypox virus antibody levels: insights from human immunological research.Virol J. 2025 May 31;22(1):175. doi: 10.1186/s12985-025-02748-0. Virol J. 2025. PMID: 40450351 Free PMC article. Review.
References
-
- Henderson, D. A. The eradication of smallpox–an overview of the past, present, and future. Vaccine29, D7–D9 (2011). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

