Comprehensive multi-omics analysis reveals the core role of glycerophospholipid metabolism in the influence of short-chain fatty acids on the development of sepsis
- PMID: 40783504
- PMCID: PMC12335468
- DOI: 10.1038/s41598-025-13322-7
Comprehensive multi-omics analysis reveals the core role of glycerophospholipid metabolism in the influence of short-chain fatty acids on the development of sepsis
Abstract
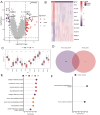
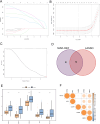
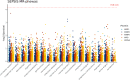
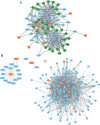
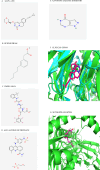
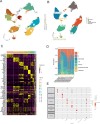
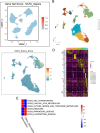
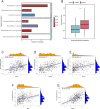
Sepsis is a systemic inflammatory response syndrome caused by infection, which has a high morbidity and mortality. Short-chain fatty acids (SCFAs) have been proved to improve the outcome of sepsis by regulating immunity and metabolism, but its specific mechanism is not clear. This study employed a multi-omics strategy integrating murine models, untargeted metabolomics, human transcriptomics (GSE185263, GSE54514), single-cell RNA sequencing (GSE167363), and Mendelian randomization to investigate SCFAs' role in sepsis. Cecal ligation and puncture (CLP) was performed in C57BL/6 mice (n = 60). Transcriptomic analysis identified 76 differentially expressed genes between septic and healthy subjects. Machine learning (SVM-RFE and LASSO regression) prioritized five SCFA-associated hub genes (CASP5, GPR84, MMP9, MPO, PRTN3), with molecular docking revealing two potential modulators. Single-cell profiling localized these targets to monocytes, while immune infiltration analysis confirmed SCFA-mediated immunomodulation. Murine metabolomics identified glycerophospholipid (GPL) metabolism as the most significantly altered pathway under SCFAs intervention. Mendelian randomization established causal relationships between GPL pathway genes and sepsis incidence/28-day mortality. Collectively, the study provide novel mechanistic and translational insights into the therapeutic targeting of short-chain fatty acids in sepsis.
Keywords: Glycerophospholipid metabolism; Multi-omics; Sepsis; Short chain fatty acids.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval and consent to participate: The following information was supplied relating to ethical approvals (i.e., approving body and any reference numbers): Ethics Committee of Guizhou Provincial People’s Hospital (Approval NO. EC Review 2023–003). Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
Integrated multi-omics analysis and predictive modeling of heart failure using sepsis-related gene signature.PLoS One. 2025 Jun 17;20(6):e0326212. doi: 10.1371/journal.pone.0326212. eCollection 2025. PLoS One. 2025. PMID: 40526611 Free PMC article.
-
Comprehensive characterization of multi-omics landscapes between gut microbial metabolites and the druggable genome in sepsis.Front Immunol. 2025 Jul 21;16:1597676. doi: 10.3389/fimmu.2025.1597676. eCollection 2025. Front Immunol. 2025. PMID: 40761792 Free PMC article.
-
Identification and Mechanistic Studies of Key Genes in Thalamic Hemorrhage Pain by Multi-omics.J Integr Neurosci. 2025 Jul 25;24(7):38130. doi: 10.31083/JIN38130. J Integr Neurosci. 2025. PMID: 40767016
-
Role of the intestinal flora-immunity axis in the pathogenesis of rheumatoid arthritis-mechanisms regulating short-chain fatty acids and Th17/Treg homeostasis.Mol Biol Rep. 2025 Jun 21;52(1):617. doi: 10.1007/s11033-025-10714-w. Mol Biol Rep. 2025. PMID: 40544212 Review.
-
Effects of Dietary Fibers on Short-Chain Fatty Acids and Gut Microbiota Composition in Healthy Adults: A Systematic Review.Nutrients. 2022 Jun 21;14(13):2559. doi: 10.3390/nu14132559. Nutrients. 2022. PMID: 35807739 Free PMC article.
References
-
- Huang, N., Wei, Y. & Wang, M. Dachaihu decoction alleviates septic intestinal epithelial barrier disruption via PI3K/AKT pathway based on transcriptomics and network pharmacology. J. Ethnopharmacol.337, 118937 (2025). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

