CRISPR/Cas9-mediated editing of carotenoid biosynthesis genes alters carotenoid concentrations in kiwifruit
- PMID: 40783509
- PMCID: PMC12335140
- DOI: 10.1186/s12870-025-07112-6
CRISPR/Cas9-mediated editing of carotenoid biosynthesis genes alters carotenoid concentrations in kiwifruit
Abstract
Background: CRISPR/Cas9 technology has garnered increasing attention for its simplicity and precision in genome editing, making it an indispensable tool for gene function research and crop genetic improvement. However, the inefficiency and time-consuming nature of genetic transformation continue to pose substantial challenges to its widespread application in woody plants.
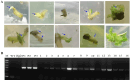
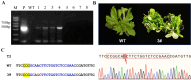
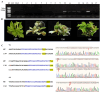
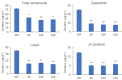
Results: In this study, we developed a rapid and efficient Agrobacterium-mediated transformation system using petioles as explants for kiwifruit. Positive resistant seedlings were obtained within three months by inoculating on MS medium supplemented with 2.0 mg·L-1 6-benzylaminopurine (6-BA), 0.2 mg·L-1 naphthaleneacetic acid (NAA), and 10 mg·L-1 hygromycin, which was faster than using leaves as explants. Using this system, CRISPR/Cas9-mediated editing of phytoene desaturase (AcPDS) and ζ-carotene desaturase (AcZDS) achieved an editing efficiency of 20%. Transgenic kiwifruit lines with edited AcZDS exhibited a significant reduction in carotenoid content.
Conclusions: Overall, we established an efficient Agrobacterium-mediated transformation system using petioles as explants, which is applicable for CRISPR/Cas9-mediated gene editing in kiwifruit, thereby facilitating functional gene studies and genetic improvement.
Keywords: ζ-carotene desaturase (ZDS); CRISPR/Cas9; Genetic transformation; Kiwifruit; Phytoene desaturase (PDS).
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
Multiplex CRISPR-Cas9 editing of chlorophyll biosynthesis genes in chickpea via protoplast and Agrobacterium-mediated transformation.Funct Integr Genomics. 2025 Jul 29;25(1):163. doi: 10.1007/s10142-025-01665-3. Funct Integr Genomics. 2025. PMID: 40728780
-
CRISPR/Cas genome editing in soybean: challenges and new insights to overcome existing bottlenecks.J Adv Res. 2025 Jul;73:53-72. doi: 10.1016/j.jare.2024.08.024. Epub 2024 Aug 18. J Adv Res. 2025. PMID: 39163906 Free PMC article. Review.
-
Efficient and versatile rapeseed transformation for new breeding technologies.Plant J. 2025 Jul;123(1):e70330. doi: 10.1111/tpj.70330. Plant J. 2025. PMID: 40639408 Free PMC article.
-
Agrobacterium-mediated transformation for virus-induced gene silencing (VIGS) in Castilleja tenuiflora: Cte-chlH and Cte-PDS.Plant Cell Rep. 2025 Jul 29;44(8):183. doi: 10.1007/s00299-025-03566-y. Plant Cell Rep. 2025. PMID: 40730887
-
Empowering Agrobacterium: Ternary vector systems as a new arsenal for plant transformation and genome editing.Biotechnol Adv. 2025 Oct;83:108631. doi: 10.1016/j.biotechadv.2025.108631. Epub 2025 Jun 24. Biotechnol Adv. 2025. PMID: 40571168 Review.
References
-
- Jiao R, Gao C. The CRISPR/Cas9 genome editing revolution. J Genet Genomics. 2016;43(5):227–8. 10.1016/j.jgg.2016.05.004. - PubMed
-
- Liu W, Zhu X, Lei M, Xia Q, Botella JR, Zhu JK, Mao Y. A detailed procedure for CRISPR/Cas9 mediated gene editing in Arabidopsis thaliana. Sci Bull. 2015;60(15):1332–47. 10.1007/s11434-015-0848-2.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

