Cell free RNA detection of pancreatic cancer in pre diagnostic high risk and symptomatic patients
- PMID: 40783559
- PMCID: PMC12335587
- DOI: 10.1038/s41467-025-62685-y
Cell free RNA detection of pancreatic cancer in pre diagnostic high risk and symptomatic patients
Abstract
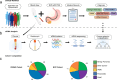
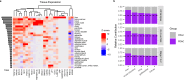
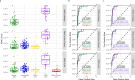
Pancreatic ductal adenocarcinomas (PDAC) are among the most fatal cancers, in part due to frequent detection at advanced stages. Endoscopic ultrasound-guided fine-needle aspiration (EUS-FNA), the most sensitive diagnostic method of PDAC in current standard clinical practice, is invasive, costly, with access limited to major healthcare settings. Here, we present a non-invasive evaluation of plasma cell-free RNA (cfRNA) for PDAC detection in pre-diagnostic high-risk and de novo symptomatic patients presenting for EUS-FNA. We develop a cfRNA normalization method to account for preanalytical variation and handling effects and derive 29 potential cfRNA biomarkers for PDAC diagnosis using 153 samples collected prior to the EUS procedure. Biomarkers related to liver function are elevated in PDAC samples, including early-stage patients without liver metastasis. Classification of PDAC using these biomarkers is validated using an independent cohort of 95 samples. Our findings could help to improve diagnostic utility in high-risk and symptomatic individuals.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

