Multi-omics insights of immune cells in the risk and prognosis of idiopathic membranous nephropathy
- PMID: 40783650
- PMCID: PMC12335521
- DOI: 10.1038/s42003-025-08642-3
Multi-omics insights of immune cells in the risk and prognosis of idiopathic membranous nephropathy
Abstract
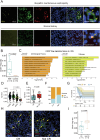
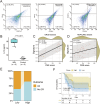
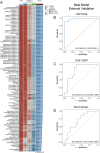
Idiopathic membranous nephropathy (IMN) is the major cause of autoimmune-related nephrotic syndrome. The role immune cells play in the risk and prognosis of IMN remains elusive. We employ multi-omics data and a variety of approaches to evaluate the causal link between 731 immune-cell phenotypes and IMN. In light of the findings emanating from Mendelian randomization analyses, only the regulatory T cell (Treg) subtype (CD39+ Tregs) survived from Bonferroni correction and is causally related to IMN. These cells are significantly enriched in the IMN microenvironment and are negatively correlated with treatment response and prognosis. We validate our findings through multiple immunofluorescence staining and explore the characteristics of CD39+ Tregs using Single-cell transcriptome analysis and flow cytometry. Based on the signature genes of CD39+ Tregs, we construct 107 composited machine-learning models to identify MN. We show the substantial contribution of CD39+ Tregs in both the risk factor determination and prognosis of IMN.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
Phenotypical analysis of ectoenzymes CD39/CD73 and adenosine receptor 2A in CD4+ CD25high Foxp3+ regulatory T-cells in psoriasis.Australas J Dermatol. 2018 Feb;59(1):e31-e38. doi: 10.1111/ajd.12561. Epub 2017 Mar 15. Australas J Dermatol. 2018. PMID: 28295154 Free PMC article.
-
Immunosuppressive treatment for idiopathic membranous nephropathy in adults with nephrotic syndrome.Cochrane Database Syst Rev. 2014 Oct 16;2014(10):CD004293. doi: 10.1002/14651858.CD004293.pub3. Cochrane Database Syst Rev. 2014. Update in: Cochrane Database Syst Rev. 2021 Nov 15;11:CD004293. doi: 10.1002/14651858.CD004293.pub4. PMID: 25318831 Free PMC article. Updated.
-
Antisense to human CD39 dysregulates immune metabolism in inflammatory bowel disease.Cell Mol Immunol. 2025 Jul;22(7):730-742. doi: 10.1038/s41423-025-01295-6. Epub 2025 May 26. Cell Mol Immunol. 2025. PMID: 40415115 Free PMC article.
-
An update on clinical significance of use of THSD7A in diagnosing idiopathic membranous nephropathy: a systematic review and meta-analysis of THSD7A in IMN.Ren Fail. 2018 Nov;40(1):306-313. doi: 10.1080/0886022X.2018.1456457. Ren Fail. 2018. PMID: 29623759 Free PMC article.
-
CD39 delineates chimeric antigen receptor regulatory T cell subsets with distinct cytotoxic & regulatory functions against human islets.Front Immunol. 2024 Jun 28;15:1415102. doi: 10.3389/fimmu.2024.1415102. eCollection 2024. Front Immunol. 2024. PMID: 39007132 Free PMC article.
References
-
- Ronco, P. et al. Membranous nephropathy. Nat. Rev. Dis. Prim.7, 69 (2021). - PubMed
-
- Mcgrogan, A., Franssen, C. F. & De Vries, C. S. The incidence of primary glomerulonephritis worldwide: a systematic review of the literature. Nephrol. Dial. Transpl.26, 414–430 (2011). - PubMed
-
- Cattran, D. C. & Brenchley, P. E. Membranous nephropathy: integrating basic science into improved clinical management. Kidney Int91, 566–574 (2017). - PubMed
-
- Ronco, P. & Debiec, H. Pathophysiological advances in membranous nephropathy: time for a shift in patient’s care. Lancet385, 1983–1992 (2015). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

