SPINT1-AS1 promotes oxidative damage and apoptosis of gastric cancer cells via the miR-656-3p/PLCXD3 axis
- PMID: 40783674
- PMCID: PMC12335420
- DOI: 10.1007/s12672-025-03195-7
SPINT1-AS1 promotes oxidative damage and apoptosis of gastric cancer cells via the miR-656-3p/PLCXD3 axis
Abstract
Background and objectives: In the study of gastric cancer (GC), long non-coding RNAs (lncRNAs) have been identified and their functions have been partly characterized; however, the specific function of SPINT1 antisense RNA 1 (SPINT1-AS1) in GC remains unclear. This study aimed to investigate the role of SPINT1-AS1 in GC cells and elucidate its downstream molecular mechanisms.
Methods: SPINT1-AS1, microRNA-656-3p (miR-656-3p), and phospholipase C, X domain containing 3 (PLCXD3) levels were modulated in GC cells through transfection experiments. Quantitative reverse transcription polymerase chain reaction and Western blot analyses were performed to assess SPINT1-AS1, miR-656-3p, and PLCXD3 levels. The proliferative capacity, apoptosis, invasion, migration, and oxidative stress levels of GC cells were evaluated using 5-ethynyl-2'-deoxyuridine assay, colony formation assay, flow cytometry, Transwell assays, and commercial kits, respectively. Dual-luciferase reporter assay and RNA immunoprecipitation assay were conducted to assess the targeting relationships among SPINT1-AS1, miR-656-3p, and PLCXD3. The impact of SPINT1-AS1 on GC tumor growth was examined in xenograft tumor models.
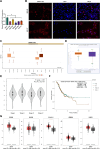
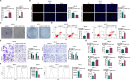
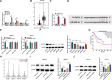
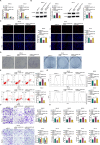
Results: SPINT1-AS1 and PLCXD3 were found to be downregulated in GC, whereas miR-656-3p was upregulated. SPINT1-AS1 elevation inhibited GC cell proliferation, invasion, migration, and oxidative stress, and promoted apoptosis. SPINT1-AS1 knockdown had opposite effects. The pro-tumor effects induced by SPINT1-AS1 knockdown were reversed by concomitant knockdown of miR-656-3p. Similarly, the inhibitory effects of SPINT1-AS1 elevation on GC malignancy were abrogated by PLCXD3 knockdown. SPINT1-AS1 knockdown suppressed GC tumor growth in mice. SPINT1-AS1 competitively bound to miR-656-3p to mediate PLCXD3 expression.
Conclusions: SPINT1-AS1 suppresses GC malignancy through the regulation of the miR-656-3p/PLCXD3 axis. These findings provide robust data supporting the biological functions of lncRNAs in GC and offer potential targets for therapeutic intervention.
Keywords: Gastric cancer; Oxidative damage; PLCXD3; SPINT1-AS1; miR-656-3p.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval and consent to participate: All animal experiments were complied with the ARRIVE guidelines and performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The experiments were approved by the Institutional Animal Care and Use Committee of Shaanxi Provincial Cancer Hospital (No. 2020-XA024). Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
LRRC75A-AS1 facilitates breast cancer cell proliferation and invasion via functioning as a CeRNA to modulate miR489-3p/ARD1.Sci Rep. 2025 Aug 26;15(1):31501. doi: 10.1038/s41598-025-17372-9. Sci Rep. 2025. PMID: 40858914 Free PMC article.
-
Long noncoding RNA MATN1-AS1 contributes to oxaliplatin resistance of gastric cancer cells through miR-518b/ZNF281 axis.Naunyn Schmiedebergs Arch Pharmacol. 2025 Aug;398(8):11041-11060. doi: 10.1007/s00210-025-03990-7. Epub 2025 Mar 12. Naunyn Schmiedebergs Arch Pharmacol. 2025. PMID: 40072551
-
LncRNA PCED1B-AS1 mediates miR-3681-3p/MAP2K7 axis to promote metastasis, invasion and EMT in gastric cancer.Biol Direct. 2024 May 2;19(1):34. doi: 10.1186/s13062-024-00468-z. Biol Direct. 2024. PMID: 38698487 Free PMC article.
-
A Novel circ_0104345/miR-876-3p/ZBTB20 Axis Regulates the Proliferation, Migration, Invasion, and Apoptosis of Breast Cancer Cells.Biochem Genet. 2023 Dec;61(6):2548-2565. doi: 10.1007/s10528-023-10391-z. Epub 2023 May 6. Biochem Genet. 2023. PMID: 37148331
-
Regulation of Human Lung Adenocarcinoma Cell Proliferation by LncRNA AFAP-AS1 Through the miR-508/ZWINT Axis.Int J Mol Sci. 2025 Jul 7;26(13):6532. doi: 10.3390/ijms26136532. Int J Mol Sci. 2025. PMID: 40650307 Free PMC article.
References
-
- Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396(10251):635–48. - PubMed
-
- Hall CE, Maegawa F, Patel AD, Lin E. Management of gastric cancer. Am Surg. 2023;31348221148359.
-
- Gill JG, Piskounova E, Morrison SJ, Cancer. Oxidative stress, and metastasis. Cold Spring Harb Symp Quant Biol. 2016;81:163–75. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
