RNA-binding proteins regulate immune-related alternative splicing in inherited salt-losing tubulopathies
- PMID: 40783707
- PMCID: PMC12335119
- DOI: 10.1186/s13023-025-03972-1
RNA-binding proteins regulate immune-related alternative splicing in inherited salt-losing tubulopathies
Abstract
Background: Inherited salt-losing tubulopathies (SLT) are rare disorders caused by gene mutations that disrupt renal tubular ion transport. However, the molecular mechanisms underlying SLT pathogenesis remain unclear. This study aims to elucidate the functional genes and potential regulatory mechanisms associated with SLT.
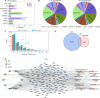
Methods: We established a study cohort comprising inherited SLT patients, age-matched patients with acquired hypokalemia, and healthy volunteers. Clinical characteristics were compared among the groups. RNA sequencing (RNA-seq) was performed to obtain transcriptomic profiles, followed by analysis of gene expression patterns and alternative splicing events (ASEs). Key findings were validated using RT-qPCR.
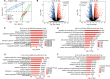
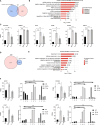
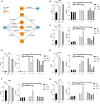
Results: SLT patients exhibited a higher prevalence of recurrent viral infections (65%, P = 0.004) and autoimmune thyroid disorders (30%, P = 0.022) compared to healthy controls. RNA-seq analysis identified 2,611 differentially expressed genes (DEGs) in SLT patients, including 1,236 upregulated and 1,375 downregulated genes. These DEGs were primarily enriched in innate immune responses and adaptive immunity pathways. Additionally, significant alterations in gene expression related to viral defense and stress responses were observed. Notably, we identified several RNA-binding proteins (RBPs) that may contribute to SLT pathogenesis by regulating ASEs of immune-related genes.
Conclusion: Our findings highlight the critical role of RBPs in SLT pathogenesis and provide novel insights into the immune profiles and gene expression dynamics in SLT. This study lays the foundation for future research into targeted therapies and personalized treatment strategies for SLT management.
Keywords: Alternative splicing events; Hypokalemia; Immune; Inherited salt-losing tubulopathies; RNA-binding proteins; Transcriptomic.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study was approved by the Ethics Committee of the People’s Hospital of Xinjiang Uygur Autonomous Region in China (No.KY2021052650). Consent for publication: Informed consent has been obtained from all subjects or their parents. Competing interests: The authors declare that they have no competing interests.
Figures

Similar articles
-
Involvement of dysregulated RNA binding protein and alternative splicing regulatory networks in diabetic nephropathy from type 2 albuminuric cohorts.BMC Nephrol. 2025 Jul 1;26(1):326. doi: 10.1186/s12882-025-04237-6. BMC Nephrol. 2025. PMID: 40597902 Free PMC article.
-
ALDH18A1 has carcinogenic functions and regulates alternative splicing events of DNA repair-related genes in esophageal carcinoma cells.Sci Rep. 2025 Aug 7;15(1):28845. doi: 10.1038/s41598-025-08006-1. Sci Rep. 2025. PMID: 40770266 Free PMC article.
-
RBM47 functions as an anti-oncogene by regulating expression and alternative splicing of cell proliferation and apoptosis associated genes in colorectal cancer cells.Sci Rep. 2025 Jul 22;15(1):26685. doi: 10.1038/s41598-025-05151-5. Sci Rep. 2025. PMID: 40695891 Free PMC article.
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
References
-
- Simon DB, Nelson-Williams C, Bia MJ, Ellison D, Karet FE, Molina AM, et al. Gitelman’s variant of Bartter’s syndrome, inherited hypokalaemic alkalosis, is caused by mutations in the thiazide-sensitive Na-Cl cotransporter. Nat Genet. 1996;12:24–30. - PubMed
-
- Koulouridis E, Koulouridis I. Molecular pathophysiology of Bartter’s and Gitelman’s syndromes. World J Pediatr. 2015;11:113–25. - PubMed
-
- Kim YK, Song HC, Kim W-Y, Yoon HE, Choi Y-J, Ki C-S, et al. Acquired Gitelman syndrome in a patient with primary Sjögren syndrome. Am J Kidney Dis. 2008;52:1163–7. - PubMed
-
- Venkat Raman G, Albano JD, Millar JG, Lee HA. Bartter’s syndrome and diabetes mellitus. J Intern Med. 1990;228:525–31. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

