CD24a knockout results in an enhanced macrophage- and CD8⁺ T cell-mediated anti-tumor immune responses in tumor microenvironment in a murine triple-negative breast cancer model
- PMID: 40783744
- PMCID: PMC12335121
- DOI: 10.1186/s12929-025-01165-3
CD24a knockout results in an enhanced macrophage- and CD8⁺ T cell-mediated anti-tumor immune responses in tumor microenvironment in a murine triple-negative breast cancer model
Abstract
Background: CD24 plays a crucial role not only in promoting tumor progression and metastasis but also in modulating macrophage-mediated anti-tumor immunity. However, its impact on the immune landscape of the tumor microenvironment (TME) remains unexplored. Here, we investigated the role of CD24a, the murine CD24 gene, in tumor progression and TME immune dynamics in a murine triple-negative breast cancer (TNBC) model.
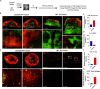
Methods: Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)/Cas9 knockout technology was employed to generate CD24a knockout in the murine TNBC cell line 4T1. Flow cytometry was utilized to analyze the immune cell populations, including myeloid-derived suppressor cells (MDSCs), natural killer cells, T cells, and macrophages, within tumors, spleens, and bone marrow in the orthotopic mouse 4T1 breast cancer model. Immunofluorescence (IF) staining was used to detect the immune cells in tumor sections. High-speed confocal was used to perform three-dimensional (3D) mapping of immune cells in the 4T1 orthotopic tumors.
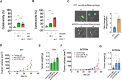
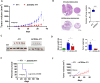
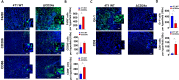
Results: Knocking out CD24a significantly reduced tumor growth kinetics and prolonged mouse survival in vivo. Flow cytometry and IF analysis of tumor samples revealed that CD24a loss significantly promoted the infiltration of M1 macrophages and cytotoxic CD8+ T cells into the TME while reducing the recruitment and expansion of granulocytic MDSCs (gMDSCs). In vitro coculture experiments showed that CD24a deficiency significantly enhanced macrophage-mediated phagocytosis and CD8⁺ T cell-mediated cytotoxicity, effects that were partially reversed by re-expression of CD24a. Moreover, in vivo depletion of macrophages and CD8+ T cells reverted the delayed tumor growth caused by CD24a knockout, underscoring their critical role in tumor growth suppression associated with CD24a knockout. 3D mapping of immune cells in the TME confirmed the anti-tumor immune landscape in the CD24a knockout 4T1 tumors. Furthermore, in vitro analysis showed that CD24a loss upregulated macrophage colony-stimulating factor expression while suppressed levels of CXCL1, CXCL5, and CXCL10, chemokines known to recruit gMDSCs, further providing a molecular basis for enhanced macrophage recruitment and diminished gMDSC accumulation.
Conclusions: Our findings suggest that CD24a may regulate immune suppression within the TNBC TME. Targeting CD24a enhances macrophage- and CD8⁺ T cell-mediated anti-tumor immune responses and is associated with a shift in the TME toward a more immunogenic state, thereby suppressing tumor growth. These results may support CD24 as a promising immunotherapeutic target for TNBC.
Keywords: CD24a; CD8+ T cells; CRISPR/Cas9 knockout; Macrophages; Triple-negative breast cancer; Tumor microenvironment.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All animal experiments received approval from the Animal Use Protocol Committee at China Medical University (CMUIACUC number: 2022-362) and were conducted in compliance with the National Institutes of Health (NIH) guidelines for the Care and Use of Laboratory Animals. Consent for publication: Not applicable. Competing interests: The authors have stated that they have no competing interests.
Figures



Similar articles
-
Salvianic acid A enhances anti-PD-1 therapy by promoting HEV-mediated stem-like CD8 T cells infiltration in TNBC.Cancer Immunol Immunother. 2025 Jun 30;74(8):256. doi: 10.1007/s00262-025-04116-x. Cancer Immunol Immunother. 2025. PMID: 40586931 Free PMC article.
-
Interplay between tumor mutation burden and the tumor microenvironment predicts the prognosis of pan-cancer anti-PD-1/PD-L1 therapy.Front Immunol. 2025 Jul 24;16:1557461. doi: 10.3389/fimmu.2025.1557461. eCollection 2025. Front Immunol. 2025. PMID: 40777041 Free PMC article.
-
Genetically engineered macrophages reverse the immunosuppressive tumor microenvironment and improve immunotherapeutic efficacy in TNBC.Mol Ther. 2025 Jul 2;33(7):3339-3359. doi: 10.1016/j.ymthe.2025.03.024. Epub 2025 Mar 20. Mol Ther. 2025. PMID: 40119517
-
Triple negative breast cancer - prognostic role of immune-related factors: a systematic review.Acta Oncol. 2018 Jan;57(1):74-82. doi: 10.1080/0284186X.2017.1400180. Epub 2017 Nov 23. Acta Oncol. 2018. PMID: 29168430
-
The role of tumor microenvironment and immune cell crosstalk in triple-negative breast cancer (TNBC): Emerging therapeutic opportunities.Cancer Lett. 2025 Sep 28;628:217865. doi: 10.1016/j.canlet.2025.217865. Epub 2025 Jun 15. Cancer Lett. 2025. PMID: 40516902 Review.
References
-
- Kristiansen G, Sammar M, Altevogt P. Tumour biological aspects of CD24, a mucin-like adhesion molecule. J Mol Histol. 2004;35(3):255–62. - PubMed
-
- Altevogt P, Sammar M, Hüser L, Kristiansen G. Novel insights into the function of CD24: a driving force in cancer. Int J Cancer. 2021;148(3):546–59. - PubMed
-
- Carrion C, Guérin E, Gachard N, le Guyader A, Giraut S, Feuillard J. Adult bone marrow three-dimensional phenotypic landscape of B-cell differentiation. Cytometry B Clin Cytom. 2019;96(1):30–8. - PubMed
MeSH terms
Substances
Grants and funding
- NSTC 111-2622-B-039-005/National Science and Technology Council
- NSTC 112-2320-B-039-006/National Science and Technology Council
- NSTC 113-2320-B-039-018/National Science and Technology Council
- NSTC113-2634-F-039-001/National Science and Technology Council
- 113-2823-8-039-001/National Science and Technology Council
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

