Early-onset multivalvular disease caused by a missense variant in lamin A/C
- PMID: 40783787
- PMCID: PMC12398237
- DOI: 10.1016/j.xhgg.2025.100491
Early-onset multivalvular disease caused by a missense variant in lamin A/C
Abstract
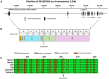
Lamins A/C, coded by LMNA gene, are crucial for nuclear architecture preservation. Pathogenic LMNA variants cause a wide range of inherited diseases called "laminopathies". A subgroup is referred to "progeroid syndromes" characterized by premature aging and other manifestations including cardiac valve abnormalities. Atypical phenotypes, generally less severe, have also been reported. We report the case of a 26-year-old male with calcific tricuspid aortic and mitral valve diseases. His father was diagnosed with severe aortic valve stenosis and mitral annulus calcification at the age of 38. The goal of this study was to identify the putative variant causing this non-syndromic multivalvular disease. Known disease-causing variants in NOTCH1, FLNA, and DCHS1 were first excluded by Sanger sequencing. Whole-exome sequencing was then performed in five family members. A LMNA variant (p.Glu262Val) was identified with in silico evidences of pathogenicity (CADD [combined annotation dependent depletion] = 33). Cells transfected with the cDNA construct harboring p.Glu262Val were characterized by abnormal nuclear morphology. Along with a literature review, the variant was classified as likely pathogenic. Elucidating the mechanism by which LMNA p.Glu262Val specifically affects cardiac heart valves is likely to provide insight about the pathogenesis of Mendelian forms of valvular heart diseases and may help guide the development of therapies.
Keywords: family study; functional study; lamin A/C; multivalvular disease; whole exome sequencing.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
-
- Thériault S., Li Z., Abner E., Luan J., Manikpurage H.D., Houessou U., Zamani P., Briend M., Estonian Biobank Research Team. Boudreau D.K., et al. Integrative genomic analyses identify candidate causal genes for calcific aortic valve stenosis involving tissue-specific regulation. Nat. Commun. 2024;15:2407. doi: 10.1038/s41467-024-46639-4. - DOI - PMC - PubMed
-
- Kyndt F., Gueffet J.-P., Probst V., Jaafar P., Legendre A., Le Bouffant F., Toquet C., Roy E., McGregor L., Lynch S.A., et al. Mutations in the gene encoding filamin A as a cause for familial cardiac valvular dystrophy. Circulation. 2007;115:40–49. doi: 10.1161/CIRCULATIONAHA.106.622621. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

