A systematic algorithm using 16S ribosomal RNA for accurate diagnosis of pneumonia pathogens
- PMID: 40784906
- PMCID: PMC12336332
- DOI: 10.1038/s41598-025-14841-z
A systematic algorithm using 16S ribosomal RNA for accurate diagnosis of pneumonia pathogens
Abstract
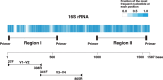
Over half of community-acquired pneumonia cases are caused by a few dozen bacterial species, and accurate identification of these pathogens is essential for effective treatment. In this study, we developed a reliable diagnostic method using 16S ribosomal RNA (16S rRNA) sequencing, considering intra-species variation, the need to differentiate Streptococcus pneumoniae from oral α-hemolytic streptococci, and applicability to the battlefield hypothesis, which helps distinguish true pathogens from commensal organisms that are not causative pathogens. We designed specific primers and a BLAST wrapper program, Cheryblast + ob, to classify 37 pneumonia-causing bacteria and 4 α-hemolytic streptococci. In simulation experiments involving a total of 20,309 copies of the 16S rRNA from 41 species of bacteria deposited in Genbank, the algorithm achieved a sensitivity greater than 0.996 and a specificity of 1.000. It was robust against sequencing errors and successfully distinguished S. pneumoniae from closely related species. In an experiment using next-generation sequencing on artificial mixtures containing genomic DNA from 10 bacterial species and human DNA at varying two-fold ratios, the species with the highest copy number was correctly identified in 8 out of 11 samples, and the top two species by copy number were identified in all 11 samples. This high-performance method offers a promising tool for accurate pneumonia diagnosis and could also be applied to other infections in which a limited number of bacterial species must be reliably identified.
Keywords: Streptococcus pneumoniae; 16S ribosomal RNA; Battlefield hypothesis; Next-generation sequencing; Pneumonia.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Performance of BIOFIRE FILMARRAY pneumonia panel in suspected pneumonia: insights from a real-world study.Microbiol Spectr. 2025 Jul;13(7):e0057125. doi: 10.1128/spectrum.00571-25. Epub 2025 May 22. Microbiol Spectr. 2025. PMID: 40401975 Free PMC article.
-
Can a Liquid Biopsy Detect Circulating Tumor DNA With Low-passage Whole-genome Sequencing in Patients With a Sarcoma? A Pilot Evaluation.Clin Orthop Relat Res. 2025 Jan 1;483(1):39-48. doi: 10.1097/CORR.0000000000003161. Epub 2024 Jun 21. Clin Orthop Relat Res. 2025. PMID: 38905450
-
What Is the Accuracy of 16S PCR Followed by Sanger Sequencing or Next-generation Sequencing in Native Vertebral Osteomyelitis? A Systematic Review and Meta-analysis.Clin Orthop Relat Res. 2025 May 1;483(5):930-938. doi: 10.1097/CORR.0000000000003314. Epub 2024 Dec 5. Clin Orthop Relat Res. 2025. PMID: 39637246
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
References
-
- American Thoracic Society; Infectious Diseases Society of America. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am. J. Respir. Crit. Care Med.171, 388–416 (2005). - PubMed
-
- Levison, M. E. & Pneumonia Including Necrotizing Pulmonary Infections (Lung Abscess). In: Harrison’s Principles of Internal Medicine, 15th Edition. McGraw-Hill, 1457–1464. (2001).
-
- Hirama, T. et al. HIRA-TAN: a real-time PCR-based system for the rapid identification of causative agents in pneumonia. Respir. Med.108, 395–404 (2014). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

