Distilling knowledge from graph neural networks trained on cell graphs to non-neural student models
- PMID: 40784939
- PMCID: PMC12336322
- DOI: 10.1038/s41598-025-13697-7
Distilling knowledge from graph neural networks trained on cell graphs to non-neural student models
Abstract
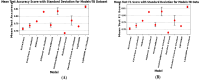
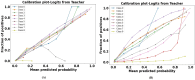
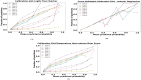
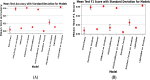
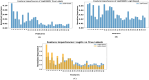
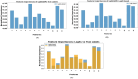
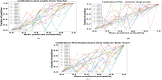
The development and refinement of artificial intelligence (AI) and machine learning algorithms have been an area of intense research in radiology and pathology, particularly for automated or computer-aided diagnosis. Whole Slide Imaging (WSI) has emerged as a promising tool for developing and utilizing such algorithms in diagnostic and experimental pathology. However, patch-wise analysis of WSIs often falls short of capturing the intricate cell-level interactions within local microenvironment. A robust alternative to address this limitation involves leveraging cell graph representations, thereby enabling a more detailed analysis of local cell interactions. These cell graphs encapsulate the local spatial arrangement of cells in histopathology images, a factor proven to have significant prognostic value. Graph Neural Networks (GNNs) can effectively utilize these spatial feature representations and other features, demonstrating promising performance across classification tasks of varying complexities. It is also feasible to distill the knowledge acquired by deep neural networks to smaller student models through knowledge distillation (KD), achieving goals such as model compression and performance enhancement. Traditional approaches for constructing cell graphs generally rely on edge thresholds defined by sparsity/density or the assumption that nearby cells interact. However, such methods may fail to capture biologically meaningful interactions. Additionally, existing works in knowledge distillation primarily focus on distilling knowledge between neural networks. We designed cell graphs with biologically informed edge thresholds or criteria to address these limitations, moving beyond density/sparsity-based definitions. Furthermore, we demonstrated that student models do not need to be neural networks. Even non-neural models can learn from a neural network teacher. We evaluated our approach across varying dataset complexities, including the presence or absence of distribution shifts, varying degrees of imbalance, and different levels of graph complexity for training GNNs. We also investigated whether softened probabilities obtained from calibrated logits offered better guidance than raw logits. Our experiments revealed that the teacher's guidance was effective when distribution shifts existed in the data. The teacher model demonstrated decent performance due to its higher complexity and ability to use cell graph structures and features. Its logits provided rich information and regularization to students, mitigating the risk of overfitting the training distribution. We also examined the differences in feature importance between student models trained with the teacher's logits and their counterparts trained on hard labels. In particular, the student model demonstrated a stronger emphasis on morphological features in the Tuberculosis (TB) dataset than the models trained with hard labels. This emphasis aligns closely with the features that pathologists typically prioritize for diagnostic purposes. Future work could explore designing alternative teacher models, evaluating the proposed approach on larger datasets, and investigating causal knowledge distillation as a potential extension.
Keywords: Cell graphs; Graph neural networks; Knowledge distillation; Non-neural models; Tuberculosis; Whole slide imaging.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures













Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
-
[Volume and health outcomes: evidence from systematic reviews and from evaluation of Italian hospital data].Epidemiol Prev. 2013 Mar-Jun;37(2-3 Suppl 2):1-100. Epidemiol Prev. 2013. PMID: 23851286 Italian.
References
-
- Yener, B. Cell-graphs: Image-driven modeling of structure-function relationship. Commun. ACM60, 74–84 (2016).
-
- World Health Organization, Global Tuberculosis Report 2024 (World Health Organization, 2024).
MeSH terms
LinkOut - more resources
Full Text Sources

