Plasmid-driven clonal expansion of multidrug-resistant monophasic Salmonella Typhimurium in a Global Food Trade Hub
- PMID: 40785095
- PMCID: PMC12340968
- DOI: 10.1080/22221751.2025.2542251
Plasmid-driven clonal expansion of multidrug-resistant monophasic Salmonella Typhimurium in a Global Food Trade Hub
Abstract
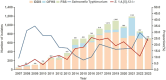
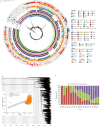
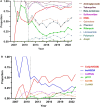
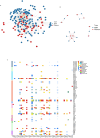
Shenzhen, a major port city with a heavily imported food supply, offers a critical setting to examine the spread and adaptation of multidrug-resistant Salmonella 1,4,[5],12:i: - (S. 1,4,[5],12:i:-). This study integrates 17 years of genomic, epidemiological, and food safety data. We explored the serovar's population structure, antibiotic resistance gene (ARG) patterns, and transmission dynamics locally and globally. Our analyses revealed substantial rise in S. 1,4,[5],12:i: - prevalence among non-typhoidal Salmonella isolates over the past 17 years, from 2.27% in 2007 to 24.79% in 2023. S. 1,4,[5],12:i: - was predominated by ST34 (97.9%), with high genotypic resistance to aminoglycosides (100%), tetracyclines (96.6%), β-lactams (89.3%), and sulphonamides (88.5%). Phylogenetic analysis separated S. 1,4,[5],12:i: - into four clades. Clade 4, first detected in Shenzhen in 2013, emerged as the predominant lineage by 2023 (56.9%). This clade exhibited minimal genetic diversity (≤ 38 core SNPs), with adaptive traits linked to the acquisition of resistance-associated plasmids. Notably, plasmid-driven ARGs, including carbapenem resistance genes, have emerged as a growing concern. Transmission analysis identified two key transmission dynamics: transient outbreaks primarily involving food handlers and persistent lineages sustained through local and international spread, often facilitated by the food supply chain. These findings underscore the role of occupational carriers and imported food products in the dissemination of ARGs, emphasizing the need for enhanced surveillance and improved health and hygiene practices for food handlers. This study provides a comprehensive molecular epidemiological framework for addressing multidrug-resistant Salmonella in globalized urban food hubs, offering a foundation for future surveillance and control efforts.
Keywords: Salmonella Typhimurium monophasic variant; food handlers; multidrug resistance (MDR); plasmid; transmission routes.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

Similar articles
-
Comprehensive analysis of β-lactam resistant non-typhoidal Salmonella Isolates: Phenotypic and genotypic insights from clinical samples in Japan.J Glob Antimicrob Resist. 2025 Jun;43:98-110. doi: 10.1016/j.jgar.2025.04.019. Epub 2025 Apr 26. J Glob Antimicrob Resist. 2025. PMID: 40294860
-
Emergence and traceability of Salmonella enterica serotype Mbandaka harboring blaOXA-10 from chickens in China.Vet Microbiol. 2025 Aug;307:110593. doi: 10.1016/j.vetmic.2025.110593. Epub 2025 Jun 9. Vet Microbiol. 2025. PMID: 40513520
-
Distinct adaptation and epidemiological success of different genotypes within Salmonella enterica serovar Dublin.Elife. 2025 Jun 25;13:RP102253. doi: 10.7554/eLife.102253. Elife. 2025. PMID: 40560760 Free PMC article.
-
A review of the global emergence of multidrug-resistant Salmonella enterica subsp. enterica Serovar Infantis.Int J Food Microbiol. 2023 Oct 16;403:110297. doi: 10.1016/j.ijfoodmicro.2023.110297. Epub 2023 Jun 22. Int J Food Microbiol. 2023. PMID: 37406596
-
The prevalence of antimicrobial drug resistance of non-typhoidal Salmonella in human infections in sub-Saharan Africa: a systematic review and meta-analysis.Expert Rev Anti Infect Ther. 2024 Sep;22(9):761-774. doi: 10.1080/14787210.2024.2368989. Epub 2024 Jun 26. Expert Rev Anti Infect Ther. 2024. PMID: 38922636
References
-
- Evolution of Salmonella enterica serotype Typhimurium driven by anthropogenic selection and niche adaptation | PLOS Genetics. https://journals.plos.org/plosgenetics/article?id = 10.1371journal.pgen.... (accessed 10 Feb2025). - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
