CFTR Gene Regulation in Human Pancreatic Duct, Bile Duct and Sweat Gland Epithelial Cells
- PMID: 40785146
- PMCID: PMC12336291
- DOI: 10.1111/jcmm.70751
CFTR Gene Regulation in Human Pancreatic Duct, Bile Duct and Sweat Gland Epithelial Cells
Abstract
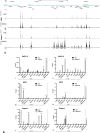
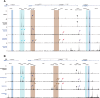
Epithelial cells at many sites in the body are affected by the inherited disorder cystic fibrosis (CF). The lung was the major focus of research until recently, when effective therapeutics became available for most people with CF. There is now renewed interest in CF aetiology in other locations in the body, where the regulatory mechanisms for the CF transmembrane conductance regulator (CFTR) gene are less well-characterised. The definition of the genomic elements controlling CFTR expression and their associated transcription factors is important for the design of gene-based therapies. Here we identify the cis-regulatory elements (CREs) associated with the CFTR locus by open chromatin mapping in pancreatic adenocarcinoma cell lines, primary human pancreatic and bile duct (cholangiocyte) organoids and single cells from tissues, as well as sweat gland coil and duct epithelial cells. We show that broadly these cell types use a combination of CREs that were characterised previously either in airway or intestinal epithelial cells, though not occurring together in these two cell lineages. Moreover, the chromatin structure of the CFTR locus in pancreatic cell lines is consistent with earlier models. We also use bioinformatic tools to predict the transcription factor network in these rare cell lineages from open chromatin peaks genome-wide.
Keywords: ATAC‐seq/snATAC‐seq; bile duct/cholangiocyte; cis‐regulatory elements; cystic fibrosis transmembrane conductance regulator gene; pancreatic duct; sweat gland.
© 2025 The Author(s). Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
K.J.G. has consulted for Genentech and received honoraria from Pfizer, holds stock in Neurocrine Biosciences, and his spouse is employed by Altos Labs Inc.
Figures




Similar articles
-
Topical cystic fibrosis transmembrane conductance regulator gene replacement for cystic fibrosis-related lung disease.Cochrane Database Syst Rev. 2016 Jun 17;2016(6):CD005599. doi: 10.1002/14651858.CD005599.pub5. Cochrane Database Syst Rev. 2016. PMID: 27314455 Free PMC article.
-
Potentiators (specific therapies for class III and IV mutations) for cystic fibrosis.Cochrane Database Syst Rev. 2015 Mar 26;(3):CD009841. doi: 10.1002/14651858.CD009841.pub2. Cochrane Database Syst Rev. 2015. Update in: Cochrane Database Syst Rev. 2019 Jan 07;1:CD009841. doi: 10.1002/14651858.CD009841.pub3. PMID: 25811419 Updated.
-
Corrector therapies (with or without potentiators) for people with cystic fibrosis with class II CFTR gene variants (most commonly F508del).Cochrane Database Syst Rev. 2023 Nov 20;11(11):CD010966. doi: 10.1002/14651858.CD010966.pub4. Cochrane Database Syst Rev. 2023. PMID: 37983082 Free PMC article.
-
Ataluren and similar compounds (specific therapies for premature termination codon class I mutations) for cystic fibrosis.Cochrane Database Syst Rev. 2023 Mar 3;3(3):CD012040. doi: 10.1002/14651858.CD012040.pub3. Cochrane Database Syst Rev. 2023. PMID: 36866921 Free PMC article.
-
A promoter-dependent upstream activator augments CFTR expression in diverse epithelial cell types.Biochim Biophys Acta Gene Regul Mech. 2024 Jun;1867(2):195031. doi: 10.1016/j.bbagrm.2024.195031. Epub 2024 Apr 27. Biochim Biophys Acta Gene Regul Mech. 2024. PMID: 38679287
References
-
- Amaral M. D. and Harrison P. T., “Development of Novel Therapeutics for All Individuals With CF (The Future Goes On),” Journal of Cystic Fibrosis 22, no. Suppl 1 (2023): S45–S49. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

