CMSCNet: a context based lightweight musculoskeletal ultrasound image segmentation method
- PMID: 40785925
- PMCID: PMC12332728
- DOI: 10.21037/qims-2024-2523
CMSCNet: a context based lightweight musculoskeletal ultrasound image segmentation method
Abstract
Background: Musculoskeletal ultrasound (MSUS) is a non-invasive and non-intrusive method for examining muscles and bones. Therefore, MSUS image analysis plays a crucial role in the assessment and early detection of musculoskeletal disorders However, due to the complexity of noise in MSUS images, analyzing and interpreting these images is a tedious and time-consuming process. Currently, ultrasound devices still rely on manual methods to analyze structural parameters such as muscle thickness, penniform angle, and fascicle length. While recent advancements in deep learning have shown promise in automating image segmentation tasks, existing methods often require extensive computational resources and may not be suitable for real-time applications. There remains a need for lightweight and efficient models that can achieve high accuracy while reducing computational load. This study aims to address this gap by proposing a context-based lightweight deep-learning framework for the automatic segmentation of MSUS images.
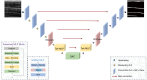
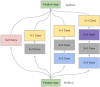
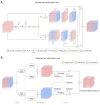
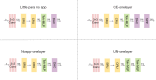
Methods: We propose a context-based lightweight deep learning framework for the automatic segmentation of MSUS images, aiming to make the segmented images as close as possible to those manually annotated by professional doctors. This method is based on the U-Net architecture, with multi-layer perception modules added to the encoder and decoder to reduce the number of parameters and improve computational efficiency. Additionally, a dense atrous convolution module is used to extract contextual features from the images, improving segmentation accuracy, and a restructured convolution module for spatial and channel dimensions is employed to reduce the extraction of redundant features during the segmentation task. We applied our method to a publicly available leg muscle dataset to extract and analyze the morphological features of the penniform muscle and conducted ablation experiments.
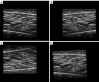
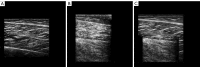
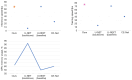
Results: The results showed that the accuracy of our algorithm was 98.7%, the same as the U-Net architecture, but with only one-tenth of the parameters. The average intersection over union (IoU) also reached 0.7227. Our network can capture more details of the muscle aponeurosis and effectively focuses on the junctions between the aponeurosis and muscle fibers, showing minimal differences from the ground truth and achieving very high accuracy.
Conclusions: This method can automatically, quickly, and accurately extract the morphological features of penniform muscles, providing a basis for improving the accuracy of muscle pathology assessment, the precision of interventional therapy, and the scientific rigor of rehabilitation treatment.
Keywords: Musculoskeletal ultrasound (MSUS); U-Net; leg muscle; segmentation.
Copyright © 2025 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-2024-2523/coif). The authors have no conflicts of interest to declare.
Figures



References
-
- Wei M, Meng D, Guo H, He S, Tian Z, Wang Z, Yang G, Wang Z. Hybrid Exercise Program for Sarcopenia in Older Adults: The Effectiveness of Explainable Artificial Intelligence-Based Clinical Assistance in Assessing Skeletal Muscle Area. Int J Environ Res Public Health 2022;19:9952. 10.3390/ijerph19169952 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
