Metagenomic Insights Into the Role of Gut Microbes in the Defensive Ink "Tsunabi" of Physeteroid Whales
- PMID: 40785991
- PMCID: PMC12334365
- DOI: 10.1002/ece3.71910
Metagenomic Insights Into the Role of Gut Microbes in the Defensive Ink "Tsunabi" of Physeteroid Whales
Abstract
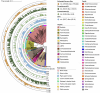
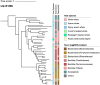
Whales of the superfamily Physeteroidea, which includes the genera Physeter and Kogia, exhibit a unique visual defense mechanism involving the release of dark reddish-brown feces (locally called "tsunabi-ink" in Japan) into the water to obscure themselves from predators and other threats. However, the mechanism underlying pigmentation remains unknown. Because physeteroids possess an enlarged distal colon that retains fecal material, a possible explanation is that symbiont microbial metabolism contributes to the feces pigmentation. To investigate this, we provided a shotgun metagenomic catalog of gut microbiomes from the intestinal tracts of eight cetacean species, including two physeteroids: a sperm whale (Physeter macrocephalus) and a pygmy sperm whale (Kogia breviceps). The colonic microbiome of physeteroids exhibited relatively high abundances of tryptophan metabolism genes, particularly indolepyruvate ferredoxin oxidoreductases (iorA and iorB), suggesting that physeteroids accumulate indole-3-pyruvate-derived pigments in their colons. Furthermore, bacterial members of the phyla Bacillota and Bacteroidota were identified in the physeteroid colon as primary taxa conferring heavy-metal resistance, which may be related to the primary predation of physeteroids on cephalopods, which bioaccumulate high levels of heavy metals. Prolonged fecal retention can expose gut microbes to chronic heavy-metal stress and colonize them as heavy metal-tolerant microbial communities, some of which may produce pigments to reduce their toxicity. Thus, we propose that tsunabi-ink is a metabolic byproduct of shifts in the gut microbial community, influenced by the host's digestive physiology and foraging behavior through sustained ecological interactions with gut symbionts. Moreover, we believe that further empirical investigation would validate this hypothesis.
Keywords: heavy metals; pigmentation; sperm whale; stranding; symbiosis; tryptophan metabolism.
© 2025 The Author(s). Ecology and Evolution published by British Ecological Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
-
- Aitchison, J. , Barceló‐Vidal C., Martín‐Fernández J. A., et al. 2000. “Logratio Analysis and Compositional Distance.” Mathematical Geology 32: 271–275. 10.1023/A:1007529726302. - DOI
Associated data
LinkOut - more resources
Full Text Sources

