Metabolism and epigenetics in cancer: toward personalized treatment
- PMID: 40786181
- PMCID: PMC12331500
- DOI: 10.3389/fendo.2025.1530578
Metabolism and epigenetics in cancer: toward personalized treatment
Abstract
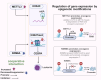
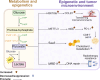
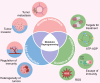
Epigenetic changes, such as DNA methylation, chromatin remodeling, and histone modifications, regulate gene expression without altering the DNA sequence. This review systematically analyzed over 500 studies including human cell line experiments (n>200), animal models (n>50), clinical cohort studies (n>100), and bioinformatics analyses retrieved from PubMed, Web of Science, and TCGA (The Cancer Genome Atlas). Studies increasingly show that genes involved in glucose and lipid metabolism, energy production, and modulation of metabolic hormones are regulated through epigenetic mechanisms. On the other hand, various metabolites participate in epigenetic modifications as coenzymes or substrates. Therefore, a greater understanding of the crosstalk between metabolism and epigenetics in cancer-related pathways could lead to the identification of key signaling molecules for targeted therapies, and raise the possibility of using dietary interventions to modulate epigenetic markers for individualized treatment. In this review, we have summarized the metabolic and epigenetic regulatory networks in cancer development, including glycolipid metabolic reprograming, the role of metabolites produced by the glut flora and tumor microenvironment, and key epigenetic drivers such as non-coding RNAs (ncRNAs). Data were curated from peer-reviewed articles, grounded in mechanistic studies using cell lines (SW480, MCF7 (Michigan cancer foundation-7)) and animal models (APC-mutant mice), with a focus on mechanistic studies, omics analyses, and translational research. Furthermore, we have discussed the potential of therapeutically targeting these pathways, along with the current challenges and future research directions, and a new strategy for reversing therapeutic drug resistance based on metabolism and epigenetic interaction was systematically explored.
Keywords: cancer; epigenetics; glucose metabolism; lipid metabolism; metabolic reprogramming; tumor microenvironment.
Copyright © 2025 Zhang, Liu, Yin, Gao, Li and Wu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Epigenetic alterations in prostate cancer: the role of chromatin remodeling.Epigenomics. 2025 Jul 22:1-25. doi: 10.1080/17501911.2025.2535938. Online ahead of print. Epigenomics. 2025. PMID: 40694614 Review.
-
[Epigenetics' implication in autism spectrum disorders: A review].Encephale. 2017 Aug;43(4):374-381. doi: 10.1016/j.encep.2016.07.007. Epub 2016 Sep 28. Encephale. 2017. PMID: 27692350 French.
-
Targeting epigenetic regulators as a promising avenue to overcome cancer therapy resistance.Signal Transduct Target Ther. 2025 Jul 18;10(1):219. doi: 10.1038/s41392-025-02266-z. Signal Transduct Target Ther. 2025. PMID: 40675967 Free PMC article. Review.
-
Epigenetic modifications in breast cancer: from immune escape mechanisms to therapeutic target discovery.Front Immunol. 2025 Apr 17;16:1584087. doi: 10.3389/fimmu.2025.1584087. eCollection 2025. Front Immunol. 2025. PMID: 40313963 Free PMC article. Review.
-
A narrative review of epigenetic marker in H3K27ac and its emerging potential as a therapeutic target in cancer.Epigenomics. 2025 Mar;17(4):263-279. doi: 10.1080/17501911.2025.2460900. Epub 2025 Feb 21. Epigenomics. 2025. PMID: 39981972 Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

