Systems metabolic engineering of Escherichia coli for the bioproduction of biliverdin and phycoerythrobilin
- PMID: 40786936
- PMCID: PMC12331624
- DOI: 10.3389/fpls.2025.1640158
Systems metabolic engineering of Escherichia coli for the bioproduction of biliverdin and phycoerythrobilin
Abstract
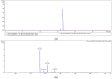
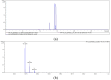
Phycobiliprotein is an important co-pigment in photosynthesis, which is composed of the covalent combination of apoprotein and phycobilin. Biliverdin IXα and phycoerythrobilin are both important substances in the phycobiliprotein biosynthesis pathway. As an economic red seaweed, Neoporphyra haitanensis has a high content of phycoerythrin. Therefore, in this study, we explored new enzyme resources for the heterologous biosynthesis of biliverdin and phycoerythrobilin. Specifically, we identified and isolated the genes encoding NhHO1, NhPebA and NhPebB from N. haitanensis, which are integral components of its phycoerythrin biosynthetic pathway. Additionally, ApHO1 from Arthrospira platensis and PebS from Prochlorococcus phage P-SSM2 were selected for comparative analysis. The results showed that genes from N. haitanensis did not encode active enzymes, which might be ascribed to the absence of crucial motifs. However, the transformation of ApHO1 and PebS into Escherichia coli could lead to the synthesis of biliverdin and phycoerythrobilin. This is the first report of sequence analysis and enzyme activity verification of phycoerythrin synthesis genes from N. haitanensis, providing a foundation for future explorations into its potential genetic resources. The successful production of biliverdin and phycoerythrobilin lay a foundation for the environmentally friendly preparation of phycobiliprotein.
Keywords: biliverdin; biosynthesis; heterologous expression; neoporphyra haitanensis; phycoerythrobilin.
Copyright © 2025 Li, Lu, Zang, Duan and Shao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Research Materials

