Long Non-Coding RNA lncTAF15:1-1 Promotes CCL5 Secretion and Cell Migration of Monocyte-Derived Dendritic Cells via PI3K/AKT/mTOR Pathway in Systemic Lupus Erythematosus
- PMID: 40787263
- PMCID: PMC12331884
- DOI: 10.2147/JIR.S527528
Long Non-Coding RNA lncTAF15:1-1 Promotes CCL5 Secretion and Cell Migration of Monocyte-Derived Dendritic Cells via PI3K/AKT/mTOR Pathway in Systemic Lupus Erythematosus
Abstract
Purpose: Systemic lupus erythematosus (SLE) is a complex autoimmune disease that seriously endangers human health. Long non-coding RNAs (lncRNAs) have been found to exhibit strong regulatory functions in cell physiology and maturation of dendritic cells (DCs). Hence, this study tried to reveal the underlying roles of one lncRNA, lncTAF15:1-1, in modulating DC functions and its involvement with CCL5 secretion in SLE pathogenesis.
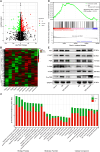
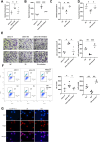
Methods: The expression levels of lncTAF15:1-1 were measured using qPCR in cultivated monocyte-derived dendritic cells (moDCs). Flow cytometry, ELISA, and Transwell chamber experiments were performed to assess various biological functions of moDCs. RNA-seq analysis was conducted to investigate transcriptional alterations in cells overexpressing lncTAF15:1-1 and negative control cells. Gene Set Enrichment Analysis (GSEA) was utilized to predict potentially involved signaling pathways, which were subsequently confirmed by Western Blotting. Rescue experiments were carried out where the expression of lncTAF15:1-1 and PI3K/AKT/mTOR pathway were altered simultaneously.
Results: LncTAF15:1-1 was significantly upregulated in moDCs from SLE patients, and it exhibited a positive correlation with SLE Disease Activity Index (SLEDAI) scores. Additionally, elevated levels of CCL5 were detected in both plasma and moDC supernatants of SLE patients. Overexpression of lncTAF15:1-1 stimulated moDCs to secrete higher levels of CCL5, and it enhanced the migration ability of moDCs as well as their capacity to attract CD4+ naïve T cells. GSEA analysis of RNA profiles indicated the potential involvement of the PI3K/AKT/mTOR pathway in lncTAF15:1-1 regulation, which was further validated by Western Blotting. The rescue experiments demonstrated that the effects of lncTAF15:1-1 on multiple functions of moDCs were attenuated when the PI3K/AKT/mTOR pathway was disrupted.
Conclusion: This study elucidated the role of lncTAF15:1-1 in orchestrating DC migration and the recruitment of CD4+ T cells by enhancing CCL5 secretion through activating PI3K/AKT/mTOR pathway, which provides insights into potential molecular targets for SLE diagnosis and treatment.
Keywords: CCL5; long non-coding RNA; monocyte-derived dendritic cells; systemic lupus erythematosus.
© 2025 Xiang et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Pharmacological effects and mechanism of Maxing Shigan Decoction in the treatment of influenza A viral pneumonia.J Ethnopharmacol. 2025 Jul 12;353(Pt A):120275. doi: 10.1016/j.jep.2025.120275. Online ahead of print. J Ethnopharmacol. 2025. PMID: 40659142
-
Upregulated SAE1 Drives Tumorigenesis and Is Associated with Poor Clinical Outcomes in Breast Cancer.Breast J. 2024 Jun 30;2024:2981722. doi: 10.1155/2024/2981722. eCollection 2024. Breast J. 2024. PMID: 39742381 Free PMC article.
-
Human Umbilical Cord Mesenchymal Stem Cells Modulate Cytokine Secretion of CD4+ T Cell in Systemic Lupus Erythematosus by Inhibiting HSP90AA1 in the Glucose-Activated PI3K-AKT Pathway.Immun Inflamm Dis. 2025 Aug;13(8):e70239. doi: 10.1002/iid3.70239. Immun Inflamm Dis. 2025. PMID: 40801323 Free PMC article.
-
Are anti-nucleosome antibodies a better diagnostic marker than anti-dsDNA antibodies for systemic lupus erythematosus? A systematic review and a study of metanalysis.Autoimmun Rev. 2012 Dec;12(2):97-106. doi: 10.1016/j.autrev.2012.07.002. Epub 2012 Jul 15. Autoimmun Rev. 2012. PMID: 22810055
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

