Development and validation of transient receptor potential channel-related signature for predicting prognosis in patients with lung adenocarcinoma
- PMID: 40787424
- PMCID: PMC12332372
- DOI: 10.3892/ol.2025.15210
Development and validation of transient receptor potential channel-related signature for predicting prognosis in patients with lung adenocarcinoma
Abstract
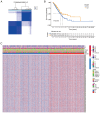
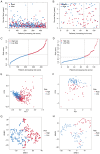
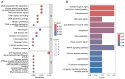
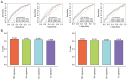
Transient receptor potential (TRP) channels are central to human temperature detection and serve a crucial role in cancer development. However, their predictive potential in patients with lung adenocarcinoma (LUAD) remains unexplored. Differential expression analysis of TRP-related genes was performed using The Cancer Genome Atlas and validated using protein-protein interaction networks. Consensus clustering stratified the LUAD subtypes. A four-gene prognostic model of anillin (ANLN), cellular repressor of E1A stimulated genes 2 (CREG2), Ras homolog family member F, filopodia associated (RHOF) and CUB domain containing protein 1 was constructed using least absolute shrinkage and selection operator-Cox regression analysis and validated in Gene Expression Omnibus and IMvigor210 cohorts. Functional enrichment, immune microenvironment analysis and drug sensitivity prediction were performed. Reverse transcription-quantitative PCR demonstrated differential expression of the prognostic genes in a LUAD cell line. A total of 43 TRP-related differentially expressed genes were identified, with consensus clustering (k=2) revealing distinct survival subgroups (P=0.015). High-risk patients exhibited immunosuppressive microenvironments with reduced B-cell infiltration and lower immunotherapy response rates (P=0.021). Functional analysis demonstrated an association of high-risk profiles with mitotic cytokinesis and chemokine signaling. A total of eight chemotherapeutic agents showed differential sensitivity between risk groups. The model outperformed existing signatures using receiver operating characteristic area under the curve and concordance-index analyses. Experimental validation demonstrated elevated expression of ANLN, CREG2 and RHOF in LUAD cells, aligning with the bioinformatics findings. The present study identified a robust TRP channel-related four-gene signature as an independent prognostic biomarker for LUAD, offering insights into immune modulation and personalized therapeutic strategies. The model's predictive accuracy for survival and treatment response underscores its translational potential in clinical decision-making.
Keywords: lung adeno-carcinoma; prognosis; signature; transient receptor potential channel.
Copyright: © 2025 He et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






Similar articles
-
Identification of different lung adenocarcinoma subtypes in combination with antidiuretic hormone-related genes and creation of an associated index to predict prognosis and guide immunotherapy.Comput Biol Chem. 2025 Dec;119:108506. doi: 10.1016/j.compbiolchem.2025.108506. Epub 2025 May 15. Comput Biol Chem. 2025. PMID: 40412340
-
Molecular subtypes of lung adenocarcinoma patients for prognosis and therapeutic response prediction with machine learning on 13 programmed cell death patterns.J Cancer Res Clin Oncol. 2023 Oct;149(13):11351-11368. doi: 10.1007/s00432-023-05000-w. Epub 2023 Jun 28. J Cancer Res Clin Oncol. 2023. PMID: 37378675 Free PMC article.
-
Unraveling the role of GPCR signaling in metabolic reprogramming and immune microenvironment of lung adenocarcinoma: a multi-omics study with experimental validation.Front Immunol. 2025 Jun 6;16:1606125. doi: 10.3389/fimmu.2025.1606125. eCollection 2025. Front Immunol. 2025. PMID: 40547013 Free PMC article.
-
Comprehensive analysis of resistance mechanisms to EGFR-TKIs and establishment and validation of prognostic model.J Cancer Res Clin Oncol. 2023 Nov;149(15):13773-13792. doi: 10.1007/s00432-023-05129-8. Epub 2023 Aug 2. J Cancer Res Clin Oncol. 2023. PMID: 37532906 Free PMC article.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
References
-
- Lung Cancer. Am Fam Physician. 2022;105:Online. No authors listed. - PubMed
-
- Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Nikšić M, Bonaventure A, Valkov M, Johnson CJ, Estève J, et al. Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): Analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391:1023–1075. doi: 10.1016/S0140-6736(17)33326-3. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
