Integrated multi-omics for potential biomarkers and molecular mechanism of persistent inflammatory refractory rheumatoid arthritis
- PMID: 40787440
- PMCID: PMC12331588
- DOI: 10.3389/fimmu.2025.1574783
Integrated multi-omics for potential biomarkers and molecular mechanism of persistent inflammatory refractory rheumatoid arthritis
Abstract
Introduction: Persistent inflammatory refractory rheumatoid arthritis (PIRRA) presents a major clinical challenge, and its underlying molecular mechanisms remain inadequately understood.
Methods: athogenesis. Synovial joint tissues were collected from 30 TgTC mice and 30 Friend virus B (FVB) control mice. Of these, 18 mice per group were used for transcriptomic, proteomic, and metabolomic analyses; 6 for pathological examination and microCT imaging; and 6 for validation experiments. Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, protein-protein interaction networks, and KEGG Markup Language (KGML) network analysis were employed to characterize the functional roles of differentially expressed genes (DEGs), proteins, metabolites, and associated biological pathways. Notably, five genes/proteins-macrophage-expressed gene 1 (Mpeg1), ectonucleotide pyrophosphatase/phosphodiesterase 2 (Enpp2), toll-like receptor 2 (Tlr2), cluster of differentiation 14 (CD14), and lysozyme 2 (Lyz2)-were validated by quantitative reverse transcription PCR (qRT-PCR), Western blotting, and immunohistochemistry.
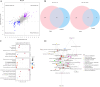
Results: A total of 2,410 DEGs, 366 differentially expressed proteins, and 120 significantly altered metabolites (P < 0.05) were identified between the model (TgTC ) and control (FVB) groups. These molecules were mainly associated with Golgi apparatus dysfunction, lipid metabolism, and immune-inflammatory responses. Integrative multi-omics analysis further revealed that these molecular alterations are involved in the activation of the PI3K-AKT-mTOR signaling pathway, as well as disruptions in tryptophan and lipid metabolism. Among the metabolites, phosphatidylinositol (PI) (12:0/12:0), N-docosahexaenoyl tryptophan, and PI (22:1(11Z)/0:0) were identified as key metabolic signatures of persistent joint synovitis in TgTC mice. In addition, the expression of Mpeg1, Enpp2, Tlr2, CD14, and Lyz2 was evaluated in synovial samples from patients with PIRRA and classical RA. Notably, Mpeg1, Enpp2, and Lyz2 were significantly upregulated in PIRRA, whereas Tlr2 and CD14 did not show statistically significant differences between groups.
Discussion: Our findings highlight the critical role of altered gene, protein, and metabolite expression in the pathogenesis of PIRRA, offering new insights into its molecular basis and potential therapeutic targets.
Keywords: golgi apparatus; lipid metabolism; multi-omics; persistent inflammatory refractory rheumatoid arthritis; tryptophan metabolism.
Copyright © 2025 Zhang, Bi, Zhao, Chen, Chen and Xiao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Rogers G, Tan Y, Shukla R, Gorodkin R, Parker B, Bruce I, et al. P209 Subgroups of refractory rheumatoid arthritis and difficult to treat features highlight differences in comorbidity and smoking history-a single-centre observational study. Rheumatology. (2022) 61:keac133.208. doi: 10.1093/rheumatology/keac133.208 - DOI
-
- Van Oosterhout M, Bajema I, Levarht E, Toes R, Huizinga T, Van Laar J. Differences in synovial tissue infiltrates between anti–cyclic citrullinated peptide–positive rheumatoid arthritis and anti–cyclic citrullinated peptide–negative rheumatoid arthritis. Arthritis Rheumatism. (2008) 58:53–60. doi: 10.1002/art.23148, PMID: - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

