Enzymatic Synthesis of an Undecorated Capsular Polysaccharide from Campylobacter jejuni
- PMID: 40788161
- PMCID: PMC12409886
- DOI: 10.1021/acs.biochem.5c00314
Enzymatic Synthesis of an Undecorated Capsular Polysaccharide from Campylobacter jejuni
Abstract
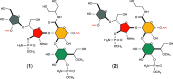
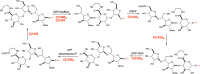
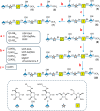
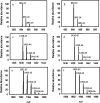
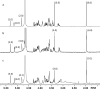
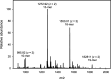
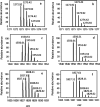
The exterior surface of the human pathogen Campylobacter jejuni is coated with a capsular polysaccharide (CPS) that helps protect it from the host immune system. In C. jejuni NCTC 11168 the repeating linear polysaccharide is composed of d-ribose, N-acetyl-d-galactosamine and d-glucuronic acid that is further amidated with either ethanolamine or serinol. The CPS is also decorated with d-glycero-l-gluco-heptose and methyl phosphoramidate. We have now shown that the polymerization of the undecorated CPS requires the sequential activity of six unique enzymes that must act in concert with one another. The catalytic activity of these six enzymes enabled a robust synthetic strategy to be developed to facilitate the assembly and isolation of specific oligosaccharides of up to 10 units in length. Modifications to this strategy enabled the isolation of mixtures containing oligosaccharides containing at least 19 monomeric units. The oligosaccharides were isolated by anion exchange chromatography and chemically characterized using ESI mass spectrometry and 1H NMR spectroscopy. These oligosaccharides will enable the reaction mechanisms for the decoration of the capsular polysaccharides to be determined and may facilitate the development of glycoconjugate vaccines.
Figures

References
-
- Tikhomirova A., McNabb E. R., Petterlin L., Bellamy G. L., Lin K. H., Santoso C. A., Daye E. S., Alhaddad F. M., Lee K. P., Roujeinikova A.. Campylobacter jejuni Virulence Factors: Update on Emerging Issues and Trends. J. Biomed. Sci. 2024;31:45. doi: 10.1186/s12929-024-01033-6. - DOI - PMC - PubMed
-
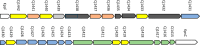
- Karlyshev A. V., Champion O. L., Churcher C., Brisson J. R., Jarrell H. C., Gilbert M., Brochu D., St Michael F., Li J. J., Wakarchuk W. W., Goodhead I., Sanders M., Stevens K., White B., Parkhill J., Wren B. W., Szymanski C. M.. Analysis of Campylobacter jejuni Capsular Loci Reveals Multiple Mechanisms for the Generation of Structural Diversity and the Ability to Form Complex Heptoses. Mol. Microbiol. 2005;55:90–103. doi: 10.1111/j.1365-2958.2004.04374.x. - DOI - PubMed
-
- Kanipes M. I., Papp-Szabo E., Guerry P., Monteiro M. A.. Mutation of waaC, Encoding Heptosyltransferase I in Campylobacter jejuni 81–176, Affects the Structure of Both Lipooligosaccharide and Capsular Carbohydrate. J. Bacteriol. 2006;188:3273–3279. doi: 10.1128/JB.188.9.3273-3279.2006. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

