Salinity Stress Induces Phase Separation of Plant BARENTSZ to Form Condensates
- PMID: 40788520
- PMCID: PMC12339837
- DOI: 10.1186/s12284-025-00830-3
Salinity Stress Induces Phase Separation of Plant BARENTSZ to Form Condensates
Abstract
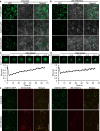
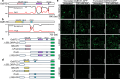
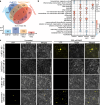
Phase separation (PS) of BARENTSZ (BTZ), a core member of the exon-junction complex (EJC), is involved in various physiological and developmental processes in animals. However, less is known about plant equivalents. Here, we demonstrated that the loss of function of Oryza sativa BTZ genes (OsBTZs) reduced rice tolerance to salinity stress. Moreover, OsBTZ proteins underwent PS independent of other core members of EJC, forming condensates under salt stress. OsBTZs may recruit proteins that play roles in the salt tolerance response to form cytoplasmic condensates, which act as stress granules. The predicted prion-like domain (PrLD), that originated ancestrally and is functionally conserved, was demonstrated to be key to the PS of OsBTZs upon NaCl treatment. This work revealed a new role for plant BTZs through an evolutionarily conserved mechanism-PS-in the formation of condensates in response to salinity stress, thus providing new insights into the adaptive evolution of plant BTZs under abiotic stress.
Keywords: BARENTSZ (BTZ); Evolution; Phase separation; Rice; Salt stress; Stress granules.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics Approval and Consent to Participate: Not applicable. Consent for Publication: Not applicable. Competing Interests: The authors declare no competing interests.
Figures


Similar articles
-
Phase separation as a key mechanism in plant development, environmental adaptation, and abiotic stress response.J Biol Chem. 2025 Jun;301(6):108548. doi: 10.1016/j.jbc.2025.108548. Epub 2025 Apr 24. J Biol Chem. 2025. PMID: 40286852 Free PMC article. Review.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Sexual Harassment and Prevention Training.2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 36508513 Free Books & Documents.
-
Boron nanoparticles combined with auxin alleviate salinity-induced oxidative stress in Oryza sativa L.Plant Sci. 2025 Oct;359:112538. doi: 10.1016/j.plantsci.2025.112538. Epub 2025 May 9. Plant Sci. 2025. PMID: 40348342
References
-
- Ali A, Petrov V, Yun DJ, Gechev T (2023) Revisiting plant salt tolerance: novel components of the SOS pathway. Trends Plant Sci 28:1060–1069 - PubMed
-
- Baguet A, Degot S, Cougot N, Bertrand E, Chenard MP, Wendling C, Kessler P, Le Hir H, Rio MC, Tomasetto C (2007) The exon-junction-complex-component metastatic lymph node 51 functions in stress-granule assembly. J Cell Sci 120:2774–2784 - PubMed
-
- Ballut L, Marchadier B, Baguet A, Tomasetto C, Séraphin B, Le Hir H (2005) The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol 12:861–869 - PubMed
-
- Bartkowska K, Tepper B, Turlejski K, Djavadian RL (2018) Roles of the exon junction complex components in the central nervous system: a mini review. Rev Neurosci 29:817–824 - PubMed
LinkOut - more resources
Full Text Sources

