Measuring the selective packaging of RNA molecules by viral coat proteins in cells
- PMID: 40789029
- PMCID: PMC12377776
- DOI: 10.1073/pnas.2505190122
Measuring the selective packaging of RNA molecules by viral coat proteins in cells
Abstract
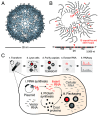
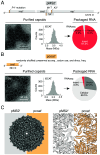
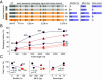
Some RNA viruses package their genomes with extraordinary selectivity, assembling protein capsids around their own viral RNA while excluding nearly all host RNA. How the assembling proteins distinguish viral RNA from host RNA is not fully understood, but RNA structure is thought to play a key role. To test this idea, we perform in-cellulo packaging experiments using bacteriophage MS2 coat proteins and a variety of RNA molecules in Escherichia coli. In each experiment, plasmid-derived RNA molecules with a specified sequence compete against the cellular transcriptome for packaging by plasmid-derived coat proteins. Following this competition, we quantify the total amount and relative composition of the packaged RNA using electron microscopy, interferometric scattering microscopy, and high-throughput sequencing. By systematically varying the input RNA sequence and measuring changes in packaging outcomes, we are able to directly test competing models of selective packaging. Our results rule out a longstanding model in which selective packaging requires the well-known translational repressor (TR) stem-loop, and instead support more recent models in which selectivity emerges from the collective interactions of multiple coat proteins and multiple stem-loops distributed across the RNA molecule. These findings establish a framework for studying and understanding selective packaging in a range of natural viruses and virus-like particles, and lay the groundwork for engineering synthetic systems that package specific RNA cargoes.
Keywords: RNA packaging; bacteriophage MS2; capsid assembly; packaging signals; plus-strand RNA virus.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures


Update of
-
Measuring the selective packaging of RNA molecules by viral coat proteins in cells.bioRxiv [Preprint]. 2025 Apr 17:2025.04.14.648219. doi: 10.1101/2025.04.14.648219. bioRxiv. 2025. Update in: Proc Natl Acad Sci U S A. 2025 Aug 19;122(33):e2505190122. doi: 10.1073/pnas.2505190122. PMID: 40321199 Free PMC article. Updated. Preprint.
Similar articles
-
Measuring the selective packaging of RNA molecules by viral coat proteins in cells.bioRxiv [Preprint]. 2025 Apr 17:2025.04.14.648219. doi: 10.1101/2025.04.14.648219. bioRxiv. 2025. Update in: Proc Natl Acad Sci U S A. 2025 Aug 19;122(33):e2505190122. doi: 10.1073/pnas.2505190122. PMID: 40321199 Free PMC article. Updated. Preprint.
-
Effect of coat-protein concentration on the self-assembly of bacteriophage MS2 capsids around RNA.Nanoscale. 2024 Feb 8;16(6):3121-3132. doi: 10.1039/d3nr03292b. Nanoscale. 2024. PMID: 38258446 Free PMC article.
-
Direct Evidence for Packaging Signal-Mediated Assembly of Bacteriophage MS2.J Mol Biol. 2016 Jan 29;428(2 Pt B):431-48. doi: 10.1016/j.jmb.2015.11.014. Epub 2015 Dec 1. J Mol Biol. 2016. PMID: 26608810 Free PMC article.
-
Factors that influence parents' and informal caregivers' views and practices regarding routine childhood vaccination: a qualitative evidence synthesis.Cochrane Database Syst Rev. 2021 Oct 27;10(10):CD013265. doi: 10.1002/14651858.CD013265.pub2. Cochrane Database Syst Rev. 2021. PMID: 34706066 Free PMC article.
-
Interventions to improve safe and effective medicines use by consumers: an overview of systematic reviews.Cochrane Database Syst Rev. 2014 Apr 29;2014(4):CD007768. doi: 10.1002/14651858.CD007768.pub3. Cochrane Database Syst Rev. 2014. PMID: 24777444 Free PMC article.
References
-
- Caspar D. L. D., Klug A., Physical principles in the construction of regular viruses. Cold Spring Harb. Symp. Quant. Biol. 27, 1–24 (1962). - PubMed
-
- Milo R., Phillips R., Cell Biology by the Numbers (Garland Science, 2015).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

