Patients-Derived Organoids Sequencing-based FOXP4 Facilitates Radioresistance by Transcriptionally Modifying GPX4 to Regulate ferroptosis in Colorectal Cancer
- PMID: 40789053
- PMCID: PMC12499432
- DOI: 10.1002/advs.202507080
Patients-Derived Organoids Sequencing-based FOXP4 Facilitates Radioresistance by Transcriptionally Modifying GPX4 to Regulate ferroptosis in Colorectal Cancer
Abstract
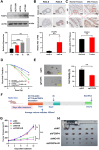
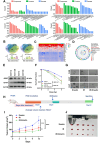
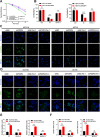
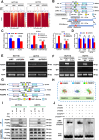
More than 50% of patients with colorectal cancer (CRC) exhibit radioresistance, indicating the need for further research on the disease. Therefore, the aim of this study is to identify radioresistance genes and elucidate the underlying molecular mechanisms using patient-derived organoids (PDOs). Transcriptome analyses are performed on radio-resistant and -sensitive PDOs, CRC cells, and xenograft tissues to screen for radioresistant genes. Additionally, the genetic homology between PDOs and clinical tissues is verified using whole-exome sequencing. Functional experiments are performed to validate the roles of the candidate genes using cellular, organoid, and animal models. Forkhead box P4 (FOXP4) is identified as a differentially expressed genes between the radio-sensitive and -resistant groups that is linked to radioresistance. Further experiments show that FOXP4 promoted radioresistance by suppressing ferroptosis. Mechanistically, FOXP4 regulated GPX4 transcription by binding to the promoter region of GPX4 via the forkhead domain to inhibit the onset of ferroptosis. Doxorubicin (DOX) inhibited FOXP4 expression by promoting its ubiquitination and degradation, eventually increasing radiosensitivity. Notably, DOX combined with irradiation attenuated the compensatory increase in FOXP4 expression and increased radiotherapy efficacy. Conclusively, the combination therapy provides a new strategy for enhancing therapeutic efficacy in CRC.
Keywords: FOXP4; Ferroptosis; GPX4; PDOs; Radioresistance.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Bray F., Laversanne M., Sung H., Ferlay J., Siegel R. L., Soerjomataram I., Jemal A., CA Cancer J. Clin. 2024, 74, 229. - PubMed
-
- Sauer R., Becker H., Hohenberger W., Rödel C., Wittekind C., Fietkau R., Martus P., Tschmelitsch J., Hager E., Hess C. F., Karstens J. H., Liersch T., Schmidberger H., Raab R., N. Engl. J. Med. 2004, 351, 1731. - PubMed
-
- van Gijn W., Marijnen C. A., Nagtegaal I. D., Kranenbarg E. M., Putter H., Wiggers T., Rutten H. J., Påhlman L., Glimelius B., van de Velde C. J., Lancet Oncol. 2011, 12, 575. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
