An RNA modification prevents extended codon-anticodon interactions from facilitating +1 frameshifting
- PMID: 40789848
- PMCID: PMC12340085
- DOI: 10.1038/s41467-025-62342-4
An RNA modification prevents extended codon-anticodon interactions from facilitating +1 frameshifting
Abstract
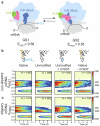
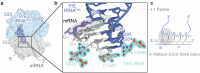
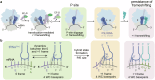
RNA post-transcriptional modifications act by stabilizing the functional conformations of RNA. While their role in messenger RNA (mRNA) decoding is well established, it is less clear how transfer RNA (tRNA) modifications outside the anticodon contribute to tRNA stability and accurate protein synthesis. Absence of such modifications causes translation errors, including mRNA frameshifting. By integrating single-molecule fluorescence resonance energy transfer and cryogenic electron microscopy, we demonstrate that the N1-methylguanosine (m1G) modification at position 37 of Escherichia coli tRNAProL is necessary and sufficient for modulating the conformational energy of this tRNA on the ribosome so as to suppress +1 frameshifting otherwise induced by this tRNA. Six structures of E. coli ribosomal complexes carrying tRNAProL lacking m1G37 show this tRNA forms four and even five codon-anticodon base pairs as it moves into the +1 frame, allowing direct visualization of the long-standing hypothesis that a four base pair codon-anticodon can form during +1 frameshifting.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
Doublet decoding of tRNASer3 demonstrates plasticity of ribosomal decoding center.Nat Commun. 2025 Jun 26;16(1):5402. doi: 10.1038/s41467-025-61016-5. Nat Commun. 2025. PMID: 40571681 Free PMC article.
-
Insights into genome recoding from the mechanism of a classic +1-frameshifting tRNA.Nat Commun. 2021 Jan 12;12(1):328. doi: 10.1038/s41467-020-20373-z. Nat Commun. 2021. PMID: 33436566 Free PMC article.
-
Genome Expansion by tRNA +1 Frameshifting at Quadruplet Codons.J Mol Biol. 2022 Apr 30;434(8):167440. doi: 10.1016/j.jmb.2021.167440. Epub 2022 Jan 4. J Mol Biol. 2022. PMID: 34995554 Free PMC article. Review.
-
Anticodon stem-loop tRNA modifications influence codon decoding and frame maintenance during translation.Semin Cell Dev Biol. 2024 Feb 15;154(Pt B):105-113. doi: 10.1016/j.semcdb.2023.06.003. Epub 2023 Jun 28. Semin Cell Dev Biol. 2024. PMID: 37385829 Free PMC article. Review.
-
Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.Nat Commun. 2021 Jul 30;12(1):4644. doi: 10.1038/s41467-021-24911-1. Nat Commun. 2021. PMID: 34330903 Free PMC article.
References
-
- Bjork, G. R., Wikstrom, P. M. & Bystrom, A. S. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science244, 986–989 (1989). - PubMed
-
- Atkins, J. F., Gesteland, R. F., Reid, B. R. & Anderson, C. W. Normal tRNAs promote ribosomal frameshifting. Cell18, 1119–1131 (1979). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

