European and African ancestry-specific plasma protein-QTL and metabolite-QTL analyses identify ancestry-specific T2D effector proteins and metabolites
- PMID: 40789849
- PMCID: PMC12339932
- DOI: 10.1038/s41467-025-62463-w
European and African ancestry-specific plasma protein-QTL and metabolite-QTL analyses identify ancestry-specific T2D effector proteins and metabolites
Abstract
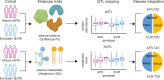
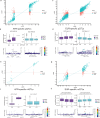
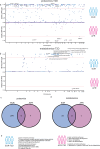
In this study, we generated and integrated plasma proteomics and metabolomics with the genotype datasets of over 2300 European (EUR) and 400 African (AFR) ancestries to identify ancestry-specific multi-omics quantitative trait loci (QTLs). In total, we mapped 954 AFR pQTLs, 2848 EUR pQTLs, 65 AFR mQTLs, and 490 EUR mQTLs. We further applied these QTLs to ancestry-stratified type-2 diabetes (T2D) risk to pinpoint key proteins and metabolites underlying the disease-associated genetic loci. Using INTACT that combined trait-imputation and colocalization results, we nominated 270 proteins and 72 metabolites from the EUR set; seven proteins and one metabolite from the AFR set as molecular effectors of T2D risk in an ancestry-stratified manner. Here, we show that the integration of genetic and omic studies of different ancestries can be used to identify distinct effector molecular traits underlying the same disease across diverse ancestral groups.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: CC has received research support from GSK and EISAI. The funders of the study had no role in the collection, analysis, or interpretation of data; in the writing of the report; or in the decision to submit the paper for publication. CC is a member of the advisory board of Circular Genomics and owns stocks. The remaining authors declare no competing interests.
Figures

Update of
-
European and African-specific plasma protein-QTL and metabolite-QTL analyses identify ancestry-specific T2D effector proteins and metabolites.Res Sq [Preprint]. 2024 Jul 22:rs.3.rs-3617016. doi: 10.21203/rs.3.rs-3617016/v1. Res Sq. 2024. Update in: Nat Commun. 2025 Aug 11;16(1):7412. doi: 10.1038/s41467-025-62463-w. PMID: 39108494 Free PMC article. Updated. Preprint.
Similar articles
-
European and African-specific plasma protein-QTL and metabolite-QTL analyses identify ancestry-specific T2D effector proteins and metabolites.Res Sq [Preprint]. 2024 Jul 22:rs.3.rs-3617016. doi: 10.21203/rs.3.rs-3617016/v1. Res Sq. 2024. Update in: Nat Commun. 2025 Aug 11;16(1):7412. doi: 10.1038/s41467-025-62463-w. PMID: 39108494 Free PMC article. Updated. Preprint.
-
Biomarker identification for Alzheimer's disease through integration of comprehensive Mendelian randomization and proteomics data.J Transl Med. 2025 Mar 6;23(1):278. doi: 10.1186/s12967-025-06317-5. J Transl Med. 2025. PMID: 40050982 Free PMC article.
-
Trans-Ancestral Genetic Risk Factors for Treatment-Related Type 2 Diabetes Mellitus in Survivors of Childhood Cancer.J Clin Oncol. 2024 Jul 1;42(19):2306-2316. doi: 10.1200/JCO.23.02281. Epub 2024 Apr 23. J Clin Oncol. 2024. PMID: 38652878 Free PMC article.
-
Population specific impact of genetic variants in KCNJ11 gene to type 2 diabetes: a case-control and meta-analysis study.PLoS One. 2014 Sep 23;9(9):e107021. doi: 10.1371/journal.pone.0107021. eCollection 2014. PLoS One. 2014. PMID: 25247988 Free PMC article.
-
Genetic risk of type 2 diabetes in populations of the African continent: A systematic review and meta-analyses.Diabetes Res Clin Pract. 2016 Apr;114:136-50. doi: 10.1016/j.diabres.2016.01.003. Epub 2016 Jan 18. Diabetes Res Clin Pract. 2016. PMID: 26830076
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

