A multichaperone condensate enhances protein folding in the endoplasmic reticulum
- PMID: 40789936
- PMCID: PMC12431857
- DOI: 10.1038/s41556-025-01730-w
A multichaperone condensate enhances protein folding in the endoplasmic reticulum
Abstract
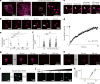
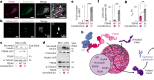
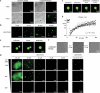
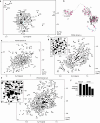
Protein folding in the endoplasmic reticulum (ER) relies on a network of molecular chaperones that facilitates the folding and maturation of client proteins. How the ER chaperones organize in a supramolecular manner to exert their cooperativity has, however, remained unclear. Here we report the discovery of a multichaperone condensate in the ER lumen, which is formed around the chaperone PDIA6 during protein folding homeostasis. The condensates form in a Ca2+-dependent manner and we resolve the underlying mechanism at the atomic and cellular levels. The PDIA6 condensates recruit further chaperones-Hsp70 BiP, J-domain protein ERdj3, disulfide isomerase PDIA1 and Hsp90 Grp94-which constitute some of the essential components of the early folding machinery. The chaperone condensates enhance folding of proteins, such as proinsulin, and prevent protein misfolding in the ER lumen. The PDIA6-scaffolded chaperone condensates hence provide the functional basis for spatial and temporal coordination of the dynamic ER chaperone network.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures







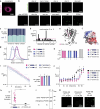


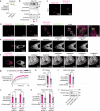
References
-
- Braakman, I. & Bulleid, N. J. Protein folding and modification in the mammalian endoplasmic reticulum. Annu. Rev. Biochem.80, 71–99 (2011). - PubMed
-
- Vincenz-Donnelly, L. & Hipp, M. S. The endoplasmic reticulum: a hub of protein quality control in health and disease. Free Radic. Biol. Med.108, 383–393 (2017). - PubMed
-
- Ellgaard, L., McCaul, N., Chatsisvili, A. & Braakman, I. Co- and post-translational protein folding in the ER. Traffic17, 615–638 (2016). - PubMed
-
- Hebert, D. N. & Molinari, M. In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol. Rev.87, 1377–13408 (2007). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

