Molecular Detection and Genetic Typing of Hepatitis E Virus in Wild Animals from Slovakia
- PMID: 40789990
- PMCID: PMC12339653
- DOI: 10.1007/s12560-025-09657-z
Molecular Detection and Genetic Typing of Hepatitis E Virus in Wild Animals from Slovakia
Abstract
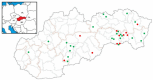
Hepatitis E is an emerging zoonosis caused by the hepatitis E virus (HEV) and is recognised worldwide. Wild boars are considered one of the main reservoirs of the zoonotic HEV-3 genotype. However, HEV-3 has also been detected in many other wildlife species. In this study, we investigated 284 liver and muscle tissue samples from wild boars and 107 liver and muscle tissue samples from four different wild ruminant species (red deer, roe deer, European mouflon and fallow deer) across 35 hunting areas in Slovakia. HEV RNA was detected in 14.2% (95% CI 9.8-18.6%) of the liver and 10.5% (95% CI 0.4-20.6%) of the muscle tissue samples from wild boars but in none of the samples from the wild ruminant species. Phylogenetic analysis based on partial ORF1 and ORF2 of the HEV genome revealed that the Slovak wild boar HEV sequences clustered within the zoonotic genotype HEV-3. Depending on their geographical origin, the obtained sequences clustered into three HEV-3 subtypes: HEV-3a, HEV-3i and HEV-3e. Our findings confirm the circulation of HEV in the wild boar population in the Slovak Republic but not in wild ruminant species.
Keywords: HEV-3 genotype; Hepatitis E virus; Wild boar; Wild ungulates; Zoonosis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Conflict of interest: The authors declare no competing interests. Research Involving Human and Animal Rights: No animal ethics approval was required for this specific study since no animals were killed for research purposes. All animals were sampled during hunting activities regulated by national and regional laws. All animals were hunted for domestic consumption.
Figures


References
-
- Beikpour, F., Borghi, M., Scoccia, E., Vicenza, T., Valiani, A., Di Pasquale, S., Bozza, S., Camilloni, B., Cozzi, L., Macellari, P., Martella, V., Suffredini, E., & Farneti, S. (2023). High prevalence and genetic heterogeneity of genotype 3 hepatitis E virus in wild boar in Umbria. Central Italy. Transboundary Emerging Diseases,2023(1), 3126419. 10.1155/2023/3126419 - DOI - PMC - PubMed
-
- Caruso, C., Modesto, P., Bertolini, S., Peletto, S., Acutis, P. L., Dondo, A., Robetto, S., Mignone, W., Orusa, R., Ru, G., & Masoero, L. (2015). Serological and virological survey of hepatitis E virus in wild boar populations in northwestern Italy: Detection of HEV subtypes 3e and 3f. Archives of Virology,160(1), 153–160. 10.1007/s00705-014-2246-5 - DOI - PubMed
-
- Caruso, C., Peletto, S., Rosamilia, A., Modesto, P., Chiavacci, L., Sona, B., Balsamelli, F., Ghisetti, V., Acutis, P. L., Pezzoni, G., Brocchi, E., Vitale, N., & Masoero, L. (2017). Hepatitis E virus: A cross-sectional serological and virological study in pigs and humans at zoonotic risk within a high-density pig farming area. Transboundary and Emerging Diseases,64(5), 1443–1453. 10.1111/tbed.12533 - DOI - PubMed
MeSH terms
Substances
Grants and funding
- VEGA 1/0220/24/The Scientific Grant Agency of the Ministry of Education, Science, Research and Sport of the Slovak Republic
- VEGA 1/0220/24/The Scientific Grant Agency of the Ministry of Education, Science, Research and Sport of the Slovak Republic
- VEGA 1/0220/24/The Scientific Grant Agency of the Ministry of Education, Science, Research and Sport of the Slovak Republic
- VEGA 1/0220/24/The Scientific Grant Agency of the Ministry of Education, Science, Research and Sport of the Slovak Republic
- VEGA 1/0220/24/The Scientific Grant Agency of the Ministry of Education, Science, Research and Sport of the Slovak Republic
- VEGA 1/0220/24/The Scientific Grant Agency of the Ministry of Education, Science, Research and Sport of the Slovak Republic
- VEGA 1/0220/24/The Scientific Grant Agency of the Ministry of Education, Science, Research and Sport of the Slovak Republic
- VEGA 1/0220/24/The Scientific Grant Agency of the Ministry of Education, Science, Research and Sport of the Slovak Republic
LinkOut - more resources
Full Text Sources

