Therapeutic application of a jumbo bacteriophage against metallo-β-lactamase producing Pseudomonas aeruginosa clinical isolates
- PMID: 40790594
- PMCID: PMC12337514
- DOI: 10.1186/s12929-025-01169-z
Therapeutic application of a jumbo bacteriophage against metallo-β-lactamase producing Pseudomonas aeruginosa clinical isolates
Abstract
Background: Therapeutic options against metallo-β-lactamase producing P. aeruginosa (MBL-PA) are limited due to multi-drug resistance. A jumbo phage isolated from wastewater in Greece was characterized microbiologically and genetically and evaluated for its potential as a therapeutic agent alone or in combination with antibiotics in an experimental thigh infection mouse model.
Methods: The host range of the jumbo phage vB_PaerM_AttikonH10 (AttikonH10) against 20 MBL-PA clinical isolates and 10 susceptible strains, one-step phage growth and growth curves of mid-exponential phase bacteria upon addition of the phage were analyzed. Whole-genome sequencing was performed and the de novo assembled complete phage genome was compared with other jumbo phages. In vivo pharmacokinetics in different tissues as well as the efficacy of two dosing regimens 109 and 106 PFU/mouse administered intraperitoneally alone and in combination with amikacin (384 mg/kg/day) was tested against an MBL-PA clinical isolate in murine thigh infection model.
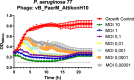
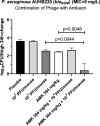
Results: The phage formed small plaques in double-layer agar and demonstrated clear or semi-clear lysis in 83.3% (25/30) of P. aeruginosa clinical isolates. Growth curves showed a > 94% growth inhibition in the presence of phage even at the lowest multiplicity of infection ratio tested (10-5). Whole genome analysis indicated a jumbo dsDNA phage with 278,406 bp (36.92% GC) belonging to Phikzvirus that is predicted to host up to 413 putative ORFs and 6 tRNA genes. No known lysogeny-associated genes, virulence factors, or antimicrobial resistance genes were identified within the genome. Phage titres 104-106 PFU/tissue were detected in all mouse tissues with elimination half-life of 3.4 h except in bronchoalveolar lavage where no phages were found. Only the high phage dose (109 PFU/mouse) reduced bacterial load in thigh by 1.09 log10 cfu/thigh compared to placebo, similar to amikacin monotherapy (1.19 log10 cfu/thigh), while their combination achieved a greater reduction of 2.07 log10 cfu/thigh compared to each monotherapy (p = 0.0044-0.0048).
Conclusions: The newly reported Phikzvirus jumbo phage AttikonH10 demonstrated a broad host range, strong lytic activity and synergistic effects with amikacin against MBL-PA isolates making it a candidate for phage therapy.
Keywords: Carbapenemase producing P. aeruginosa; Jumbo phages; Lytic activity; Metallo β-lactamases.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study has been approved by the ethics committee of the Attikon University Hospital. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures





References
-
- WHO. WHO Bacterial Priority Pathogens List 2024, 2024: bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance. Geneva: World Health Organization; 2024. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

