Hepatic CBP/p300 Orchestrate Amino Acid-Driven Gluconeogenesis through Histone Crotonylation
- PMID: 40791184
- PMCID: PMC12591208
- DOI: 10.1002/advs.202507635
Hepatic CBP/p300 Orchestrate Amino Acid-Driven Gluconeogenesis through Histone Crotonylation
Abstract
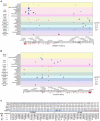
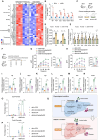
The role of amino acid metabolism dysregulation in the development of type 2 diabetes remains elusive. Here, significant associations of human CREBBP/EP300 gene polymorphisms with circulating amino acid and glucose levels are reported. Through integrated transcriptomic, metabolomic, and CUT&Tag analyses, the molecular mechanisms underlying these correlations are investigated. Liver-specific Crebbp/Ep300 double knockout mice display elevated plasma amino acid levels and impaired hepatic glucose production caused by the downregulation of amino acid metabolism genes, which is closely linked to altered histone crotonylation and acetylation patterns at their promoters. However, key gluconeogenic genes Pck1 and G6pc are not downregulated in knockout mice. Interestingly, the level of 2-aminoadipic acid (2-AAA), a biomarker of diabetes, is significantly increased due to decreased glutaryl-CoA dehydrogenase (GCDH) expression in CBP/p300-deficient livers. Treatment with 2-AAA or overexpression of GCDH enhances amino acid-driven gluconeogenesis through histone crotonylation-mediated transcriptional activation of amino acid metabolism genes in primary mouse hepatocytes, whereas GCDH knockdown exhibits an opposite result. Furthermore, targeted hepatic knockdown of CBP/p300 markedly attenuates hepatic glucose production from amino acids in diabetic mice. In sum, these findings underscore the pivotal role of CBP/p300 in linking amino acid catabolism to gluconeogenesis through histone crotonylation in a cell-autonomous manner.
Keywords: 2‐aminoadipic acid; CBP/p300; GCDH; amino acids; gluconeogenesis; histone crotonylation; type 2 diabetes.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Chandel N. S., Perspect. Biol. 2021, 13, a040584.
-
- Newgard C. B., An J., Bain J. R., Muehlbauer M. J., Stevens R. D., Lien L. F., Haqq A. M., Shah S. H., Arlotto M., Slentz C. A., Rochon J., Gallup D., Ilkayeva O., Wenner B. R., Yancy W. S., Eisenson H., Musante G., Surwit R. S., Millington D. S., Butler M. D., Svetkey L. P., Cell Metab. 2009, 9, 311. - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 2024ZD0531600/Noncommunicable Chronic Diseases-National Science and Technology Major Project
- 82270860/National Natural Science Foundation of China
- 82170819/National Natural Science Foundation of China
- 82200998/National Natural Science Foundation of China
- 82370798/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources
Miscellaneous
