This is a preprint.
Human RPL7 and DDX21 interact with HTLV-1 Gag and enhance tRNAPro primer annealing to genomic RNA
- PMID: 40791341
- PMCID: PMC12338668
- DOI: 10.1101/2025.07.15.664966
Human RPL7 and DDX21 interact with HTLV-1 Gag and enhance tRNAPro primer annealing to genomic RNA
Abstract
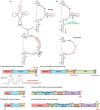
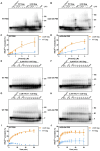
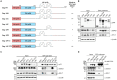
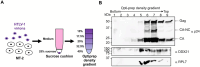
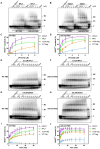
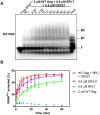
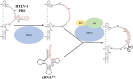
Human T-cell leukemia virus type 1 (HTLV-1), an oncogenic retrovirus, uses human tRNAPro to prime reverse transcription (RT). However, how tRNAPro is annealed to the primer binding site (PBS), which is embedded in a highly structured hairpin in the genomic RNA (gRNA), remains unclear. We hypothesize that HTLV-1 Gag may have more robust chaperone activity than mature HTLV-1 nucleocapsid (NC), which in contrast to HIV-1 NC, displays relatively weak chaperone function, and that a cellular co-factor may be required to facilitate primer tRNA annealing. Recombinant HTLV-1 Gag was successfully purified for the first time and used to perform primer-annealing assays. Relative to mature NC and matrix (MA) domains, HTLV-1 Gag is only slightly more effective at chaperoning the annealing of tRNAPro to the PBS. To identify potential HTLV-1 Gag interacting co-chaperones of tRNA annealing in cells, we performed affinity tagging/purification-mass spectrometry (AP-MS). Two significant AP-MS hits, RPL7 and DDX21, were further validated by reciprocal co-IP studies in both HEK293T and chronically HTLV-1-infected MT-2 cells. Domain mapping studies revealed that HTLV-1 Gag interacts with RPL7 and DDX21 through the zinc fingers in the NC domain independent of the presence of RNA. In addition, we showed that both RPL7 and DDX21 are packaged into virions. RPL7 or DDX21 alone was more effective than HTLV-1 Gag at annealing tRNAPro to the PBS. Synergistic effects of the Gag/RPL7/DDX21 combination in facilitating tRNAPro annealing to the PBS were found. Taken together, the mechanistic insights gained from these studies could be exploited for the development of new therapeutic strategies aimed at targeting HTLV-1 RT.
Keywords: DDX21; Gag; HTLV-1; RPL7; reverse transcription; tRNAPro primer.
Figures



References
-
- Marquet R., Isel C., Ehresmann C. and Ehresmann B. (1995) tRNAs as primer of reverse transcriptases. Biochimie, 77, 113–124. - PubMed
-
- Hargittai M.R., Gorelick R.J., Rouzina I. and Musier-Forsyth K. (2004) Mechanistic insights into the kinetics of HIV-1 nucleocapsid protein-facilitated tRNA annealing to the primer binding site. J Mol Biol, 337, 951–968. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
