This is a preprint.
Cahn-Hilliard dynamical models for condensed biomolecular systems
- PMID: 40791366
- PMCID: PMC12338720
- DOI: 10.1101/2025.07.13.664571
Cahn-Hilliard dynamical models for condensed biomolecular systems
Abstract
Biomolecular condensates create dynamic subcellular compartments that alter systems-level properties of the networks surrounding them. One model combining soluble and condensed states is the Cahn-Hilliard equation, which specifies a diffuse interface between the two phases. Customized approaches required to solve this equation are largely inaccessible. Using two complementary numerical strategies, we built stable, self-consistent Cahn-Hilliard solvers in Python, MATLAB, and Julia. The algorithms simulated the complete time evolution of condensed droplets as they dissolved or persisted, relating critical droplet size to a coefficient for the diffuse interface in the Cahn-Hilliard equation. We applied this universal relationship to the chromosomal passenger complex, a multi-protein assembly that reportedly condenses on mitotic chromosomes. The fully constrained Cahn-Hilliard simulations yielded dewetting and coarsening behaviors that closely mirrored experiments in different cell types. The Cahn-Hilliard equation tests whether condensate dynamics behave as a phase-separated liquid, and its numerical solutions advance generalized modeling of biomolecular systems.
Keywords: CPC; HeLa; MCF10A; Phase separation; cancer; multigrid; scalar auxiliary variables.
Conflict of interest statement
DECLARATION OF INTERESTS The authors declare no competing interests.
Figures
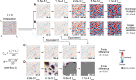
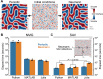




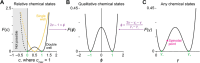
Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Electrophoresis.2025 Jul 14. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 14. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 36251838 Free Books & Documents.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Fabricating mice and dementia: opening up relations in multi-species research.In: Jenkins N, Jack-Waugh A, Ritchie L, editors. Multi-Species Dementia Studies. Bristol (UK): Bristol University Press; 2025 Feb 25. Chapter 2. In: Jenkins N, Jack-Waugh A, Ritchie L, editors. Multi-Species Dementia Studies. Bristol (UK): Bristol University Press; 2025 Feb 25. Chapter 2. PMID: 40690569 Free Books & Documents. Review.
References
-
- Yan H., Konstorum A., and Lowengrub J.S. (2018). Three-Dimensional Spatiotemporal Modeling of Colon Cancer Organoids Reveals that Multimodal Control of Stem Cell Self-Renewal is a Critical Determinant of Size and Shape in Early Stages of Tumor Growth. Bull. Math. Biol. 80, 1404–1433. 10.1007/s11538-017-0294-1. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
