This is a preprint.
SOX2 utilizes FOXA1 as a heteromeric transcriptional partner to drive proliferation in therapy-resistant prostate cancer
- PMID: 40791379
- PMCID: PMC12338689
- DOI: 10.1101/2025.07.18.664790
SOX2 utilizes FOXA1 as a heteromeric transcriptional partner to drive proliferation in therapy-resistant prostate cancer
Abstract
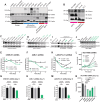
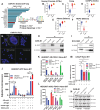
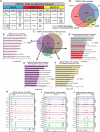
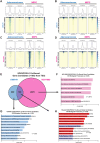
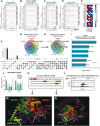
Treatment options and diagnostic outlook for men with advanced, therapy resistant prostate cancer (PCa) are extremely poor; this is primarily due to the common lack of durable response to androgen receptor (AR) targeted therapies and phenotypic transdifferentiation into a particularly lethal subtype known as neuroendocrine prostate cancer (NEPC). In this study, we mechanistically determine that SOX2 (a transcription factor originally repressed by AR) physically binds and acts in a concerted manner with FOXA1 (a key AR pioneering cofactor) to regulate a subset of genes which promote cell cycle progression, and lineage plasticity in AR-refractory prostate cancers. Our findings assert the SOX2/FOXA1 interaction as an important mediator of resistance to AR-targeted therapy and a driver of NEPC and lineage plasticity; their coordinated action and downstream signaling offers a potential novel therapeutic opportunity in late-stage PCa.
Keywords: AR; ASCL1; CRPC; FGF; FGFR; FOXA1; NEPC; Prostate cancer; ROR1; SOX2; androgen; castration; enzalutamide; lineage plasticity; neuroendocrine; stem cells.
Conflict of interest statement
The authors declare no relevant conflicts of interest related to this study. E.S.A. reports grants and personal fees from Janssen, Johnson & Johnson, Sanofi, Bayer, Bristol Myers Squibb, Convergent Therapeutics, Curium, MacroGenics, Merck, Pfizer, and AstraZeneca; personal fees from Aadi Bioscience, Abeona Therapeutics, Aikido Pharma, Astellas, Amgen, Blue Earth, Boundless Bio, Corcept Therapeutics, Duality Bio, Exact Sciences, Hookipa Pharma, Invitae, Eli Lilly, Foundation Medicine, Menarini-Silicon Biosystems, Tango Therapeutics, Tempus, Tolmar Scientific, VIR Biotechnology, and Z-alpha; grants from Novartis, Celgene, and Orion; and has a patent for an AR-V7 biomarker technology that has been licensed to Qiagen. J.M.D. has no conflicts relevant to this work. However, he serves as a consultant and Chief Scientific Officer of Astrin Biosciences. The interest related to J.M.D. has been reviewed and managed by the University of Minnesota in accordance with its Conflict-of-Interest policies.
Figures


References
-
- Sherman R.L., Firth A.U., Henley S.J., Siegel R.L., Negoita S., Sung H., Kohler B.A., Anderson R.N., Cucinelli J., Scott S., et al. (2025). Annual Report to the Nation on the Status of Cancer, featuring state-level statistics after the onset of the COVID-19 pandemic. Cancer 131, e35833. 10.1002/cncr.35833. - DOI - PMC - PubMed
-
- Isaacs J.T., Furuya Y., and Berges R. (1994). The role of androgen in the regulation of programmed cell death/apoptosis in normal and malignant prostatic tissue. Semin Cancer Biol 5, 391–400. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
