This is a preprint.
An ancient evolutionary origin for IL-1 cytokines as mediators of immunity
- PMID: 40791525
- PMCID: PMC12338702
- DOI: 10.1101/2025.07.17.662885
An ancient evolutionary origin for IL-1 cytokines as mediators of immunity
Abstract
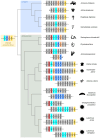
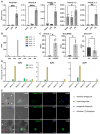
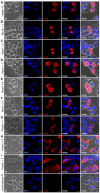
Inflammation is a hallmark of immune responses. Its mechanistic underpinnings in mammals are well-defined: pro-inflammatory cytokines of the interleukin 1 (IL-1) superfamily establish and support microenvironments that promote immune cell activities. Despite a growing number of reports on inflammatory processes and components of the IL-1 signaling axis in several invertebrate lineages, orthologs of these central cytokines have not been detected outside of the jawed vertebrates. Here, protein structure prediction algorithms were applied to identify genes encoding a family of IL-1 proteins with homologs throughout Eumetazoa (termed "IL-1anc" to reflect their ancestral evolutionary origin). Using Petromyzon marinus (sea lamprey) and Strongylocentrotus purpuratus (purple sea urchin) as model systems, we demonstrate that IL-1anc proteins share important features with mammalian IL-1α/IL-1β including expression patterns, protein localization, and processing. Together, our data indicate that the IL-1 superfamily and associated circuitry represent a foundational module of animal immunity that far predates the jawed vertebrates.
Keywords: evolution; immune response; inflammation; interleukin-1; invertebrates.
Figures



References
-
- Beck G., O’Brien R. F., Habicht G. S., Stillman D. L., Cooper E. L., and Raftos D. A. (1993). Invertebrate cytokines. III: Invertebrate interleukin-1-like molecules stimulate phagocytosis by tunicate and echinoderm cells. Cell Immunol 146, 284–299. doi: S0008-8749(83)71027-0 [pii] 10.1006/cimm.1993.1027 - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
