This is a preprint.
BLOC1S1 variants cause lysosomal and autophagic defects resulting in a hypomyelinating leukodystrophy with epileptic encephalopathy
- PMID: 40791729
- PMCID: PMC12338918
- DOI: 10.1101/2025.07.17.25331211
BLOC1S1 variants cause lysosomal and autophagic defects resulting in a hypomyelinating leukodystrophy with epileptic encephalopathy
Abstract
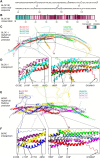
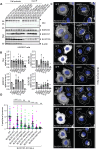
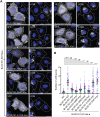
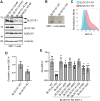
BLOC1S1 encodes a subunit shared by the BLOC-1 and BORC hetero-octameric complexes that regulate various endolysosomal processes. Here, we report the identification of seven distinct variants in BLOC1S1 in eleven individuals from seven independent families presenting with early psychomotor delay, hypotonia, spasticity, epileptic encephalopathy, optic atrophy, and leuko-axonopathy with hypomyelination. A subset of the affected individuals also have features of hypopigmentation and ocular albinism that are similar, although milder, than those of individuals with BLOC-1-related Hermansky-Pudlak syndrome. Functional analyses show that BLOC1S1 knockout (KO) impairs the anterograde transport of lysosomes and autophagy in both non-neuronal cells and iPSC-derived neurons. Rescue experiments reveal that most BLOC1S1 variants exhibit reduced expression, decreased assembly with other BORC/BLOC-1 subunits, and/or impaired restoration of lysosome transport and autophagy in BLOC1S1-KO cells. Additionally, we show that KO of BLOC1S1 reduces pigmentation in a melanocytic cell line, and that five of the BLOC1S1 variants partially or fully restore pigmentation. These findings provide genetic, clinical, and functional evidence that loss-of-function (LoF) of BLOC1S1 leads to more pronounced deficits in BORC than BLOC-1 function. We conclude that the biallelic BLOC1S1 variants characterized here primarily result in a neurological disorder with prominent leukodystrophy, similar to the recently reported condition caused by variants in the BORCS8 subunit of BORC. Together, these findings establish BORCopathies as a distinct disease entity.
Keywords: Autophagy; BLOC-1; BLOC1S1; BORC; Leukodystrophy; Lysosomes; Neurodevelopmental disorder.
Conflict of interest statement
The authors declare no competing interests. EW is CSO and equity holder of “The Organoid Company”. JLB: has grants from NIH; clinical trials with Spur Therapeutics, Calico, and Ionis; consulting with Ionis; writing content for UpToDate; stock in Orchard Therapeutics; and royalties from BioFire. EZ, AR, KT, and AMBA are employees of CENTOGENE GmbH.
Figures



References
-
- Falcón-Pérez J.M., Starcevic M., Gautam R., and Dell’Angelica E.C. (2002). BLOC-1, a novel complex containing the pallidin and muted proteins involved in the biogenesis of melanosomes and platelet-dense granules. J Biol Chem 277, 28191–28199. - PubMed
-
- Moriyama K., and Bonifacino J.S. (2002). Pallidin is a component of a multi-protein complex involved in the biogenesis of lysosome-related organelles. Traffic 3, 666–677. - PubMed
-
- Starcevic M., and Dell’Angelica E.C. (2004). Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1). J Biol Chem 279, 28393–28401. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
