Prevalence and genomic insights into type III-A CRISPR-Cas system acquisition in global Staphylococcus argenteus strains
- PMID: 40792102
- PMCID: PMC12336165
- DOI: 10.3389/fcimb.2025.1644286
Prevalence and genomic insights into type III-A CRISPR-Cas system acquisition in global Staphylococcus argenteus strains
Abstract
Introduction: The CRISPR-Cas system serves as a defense mechanism in bacteria and archaea, protecting them against the invasion of mobile genetic elements. Staphylococcus argenteus, a Gram-positive bacterium that diverged from Staphylococcus aureus, is characterized by the rare presence of the CRISPR-Cas system in only a few isolates.
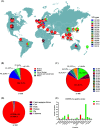
Methods: In this study, we analyzed the prevalence of the type III-A CRISPR-Cas system in 368 S. argenteus genome sequences from animals, food sources, and humans across 26 countries, available in public database.
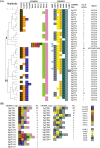
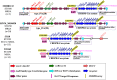
Results: Our findings revealed that 44.0% of these strains carry this immune system, with 98.1% of them belonging to the sequence type 2250 (ST2250). Genomic localization analysis indicated that the CRISPR-Cas is closely associated with SCCmec (mecA-ΔmecR1-IS1272-ccrB2-ccrA2) or Insertion sequence 1272 (IS1272) transposase. Further analysis identified a common IS1272 target inverted repeats (IR) sequence in ST2250 strains, providing insights into why these strains are more likely to acquire the CRISPR-Cas system. CRISPR typing identified 41 sequences types, classifying these strains into two clusters, with Cluster II being the predominant one. Homology analysis of spacers revealed that all the identified 15 spacers exhibited homology to sequences from plasmids, lytic phages, or prophages.
Conclusion: This study suggests that the acquisition of the CRISPR-Cas system in S. argenteus enhances its resistance to phage attacks and plasmid invasions in environmental settings, potentially posing significant challenges for clinical treatment of infections caused by these strains and hindering efforts to control their spread in food products using phage-based interventions.
Keywords: CRISPR-Cas; IS1272; SCCmec; Staphylococcus argenteus; poultry.
Copyright © 2025 Chen, Xu, Luo, Wang, Wang, Li, Jiao and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Argudín M. A., Dodémont M., Vandendriessche S., Rottiers S., Tribes C., Roisin S., et al. (2016). Low occurrence of the new species Staphylococcus argenteus in a Staphylococcus aureus collection of human isolates from Belgium. Eur. J. Clin. Microbiol. Infect. Dis. 35, 1017–1022. doi: 10.1007/s10096-016-2632-x, PMID: - DOI - PubMed
-
- Aung M. S., Osada M., Urushibara N., Kawaguchiya M., Ohashi N., Hirose M., et al. (2025). Molecular characterization of methicillin-susceptible/resistant Staphylococcus aureus from bloodstream infections in northern Japan: The dominance of CC1-MRSA-IV, the emergence of human-associated ST398 and livestock-associated CC20 and CC97 MSSA. J. Global antimicrobial resistance 41, 77–87. doi: 10.1016/j.jgar.2024.12.010, PMID: - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

