Comparative evaluation of MALDI-ToF mass spectrometry and Sanger sequencing of the 16S, hsp65, and rpoB genes for non tuberculous mycobacteria species identification
- PMID: 40792107
- PMCID: PMC12336121
- DOI: 10.3389/fcimb.2025.1612459
Comparative evaluation of MALDI-ToF mass spectrometry and Sanger sequencing of the 16S, hsp65, and rpoB genes for non tuberculous mycobacteria species identification
Abstract
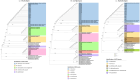
Non tuberculous mycobacteria (NTM) infections are increasing globally, underscoring the critical importance of accurate species-level identification for effective clinical management. This study aimed to evaluate the use of three conserved markers in the mycobacterial family (16S, hsp65, and rpoB) for NTM identification through Sanger sequencing, comparing the results to those obtained using MALDI-ToF MS. A total of 59 clinical NTM isolates from plastic surgery patients, previously characterized by MALDI-ToF MS, were analyzed. These isolates underwent DNA extraction, PCR amplification, and Sanger sequencing. Species identification was performed through phylogenetic analyses of each marker individually and concatenated as a multi locus sequencing approach. Concordance between MALDI-ToF MS and Sanger sequencing was assessed using Cohen's Kappa statistical analysis. Cohen's Kappa values indicated moderate concordance of 0.46 for 16S, 0.51 for hsp65, and 0.69 for rpoB. Concatenated phylogenetic analysis yielded improved concordance values of 0.71 for (16S + hsp65), 0.76 for (16S + rpoB), 0.69 for (rpoB + hsp65), and 0.72 for (16S + hsp65 + rpoB). Our results show that NTM identification is more accurate when employing a multi locus sequencing approach. Notably, the combination of 16S + rpoB outperformed the three-marker concatenation, offering the highest concordance for species-level identification. NTM identification is challenging, and concatenated phylogenetic analysis of two or more gene fragments should be used when MALDI-ToF MS or whole genome sequencing is not available.
Keywords: Ecuador; MALDI-ToF MS; Sanger sequencing; clinical performance; non-tuberculous mycobacteria.
Copyright © 2025 Rodriguez-Pazmiño, Carvajal, Paredes-Núñez, Echeverría, Calderon, Orlando, Parra Vera and Garcia-Bereguiain.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Rapid detection of rifampin resistance in Mycobacterium tuberculosis using nucleotide MALDI-TOF MS: a comparative study with phenotypic drug susceptibility testing and DNA sequencing.Microbiol Spectr. 2025 Jul;13(7):e0048325. doi: 10.1128/spectrum.00483-25. Epub 2025 May 30. Microbiol Spectr. 2025. PMID: 40445194 Free PMC article.
-
Heterogeneity among Mycobacterium avium complex species isolated from pulmonary infection in Taiwan.Microbiol Spectr. 2025 Aug 5;13(8):e0030925. doi: 10.1128/spectrum.00309-25. Epub 2025 Jul 7. Microbiol Spectr. 2025. PMID: 40621928 Free PMC article.
-
Evaluation of Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry for Identification of Nontuberculous Mycobacteria from Clinical Isolates.J Clin Microbiol. 2015 Aug;53(8):2737-40. doi: 10.1128/JCM.01380-15. Epub 2015 Jun 10. J Clin Microbiol. 2015. PMID: 26063855 Free PMC article.
-
Antibiotic treatment for non-tuberculous mycobacteria lung infection in people with cystic fibrosis.Cochrane Database Syst Rev. 2025 Mar 27;3(3):CD016039. doi: 10.1002/14651858.CD016039. Cochrane Database Syst Rev. 2025. PMID: 40145528
-
Performance of the matrix-assisted laser desorption ionization time-of-flight mass spectrometry system for rapid identification of streptococci: a review.Eur J Clin Microbiol Infect Dis. 2017 Jun;36(6):1005-1012. doi: 10.1007/s10096-016-2879-2. Epub 2017 Jan 23. Eur J Clin Microbiol Infect Dis. 2017. PMID: 28116553
References
-
- Adékambi T., Colson P., Drancourt M. (2003). rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J. Clin. Microbiol. 41, 5699–5708. doi: 10.1128/JCM.41.12.5699-5708.2003/ASSET/BE684F4A-DBD4-4FBD-9C88-969A6C9CC686/ASSETS/GRAPHIC/JM1230623003.JPEG, PMID: - DOI - PMC - PubMed
-
- Alcolea-Medina A., Fernandez M. T. C., Montiel N., García M. P. L., Sevilla C. D., North N., et al. (2019). An improved simple method for the identification of Mycobacteria by MALDI-TOF MS (Matrix-Assisted Laser Desorption- Ionization mass spectrometry). Sci. Rep. 9, 1–6. doi: 10.1038/S41598-019-56604-7;TECHMETA=140;SUBJMETA=107,2521,326,631;KWRD=CLINICAL+MICROBIOLOGY,INFECTIOUS-DISEASE+DIAGNOSTICS, PMID: - DOI - PMC - PubMed
-
- Aldous W. K., Pounder J. I., Cloud J. L., Woods G. L. (2005). Comparison of six methods of extracting Mycobacterium tuberculosis DNA from processed sputum for testing by quantitative real-time PCR. J. Clin. Microbiol. 43, 2471–2473. doi: 10.1128/JCM.43.5.2471-2473.2005/ASSET/5037D851-31C6-4C46-B2C9-CBC7272B96B8/ASSETS/GRAPHIC/ZJM0050553860001.JPEG, PMID: - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

