TSPAN9 inhibits malignant progression of hepatocellular carcinoma
- PMID: 40792132
- PMCID: PMC12335697
- DOI: 10.21037/tcr-2025-174
TSPAN9 inhibits malignant progression of hepatocellular carcinoma
Abstract
Background: Tetraspanins (TSPANs) are a critical family for cell migration, which have been implicated in a variety of activities including cancer. Previous study showed that tetraspanin 9 (TSPAN9) plays an important role in gastric cancer. In this study, we aim to explore the biological functions of TSPAN9 in hepatocellular carcinoma (HCC).
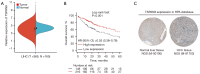
Methods: The Cancer Genome Atlas (TCGA), Genotype-Tissue Expression (GTEx) and Gene Expression Profiling Interactive Analysis (GEPIA) databases were used to analyze TSPAN9 expression, prognostic mutations, somatic copy number alterations (sCNAs), and tumor immune characteristics in 33 tumors. Immunohistochemistry images of TSPAN9 in HCC tissues were obtained from the Human Protein Atlas (HPA) database. Survival curves were generated using the Kaplan-Meier Plotter database. The proliferation, migration, and invasion abilities of HCC cells were assessed using Cell Counting Kit-8 (CCK-8), wound healing, and Transwell assays.
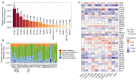
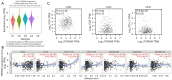
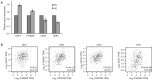
Results: TSPAN9 was deregulated in various tumor types, and its expression was lower in HCC tissues than that of normal liver tissues (P<0.05). Moreover, TSPAN9 expression had a correlation with the prognosis of tumor patients, and HCC patients with low TSPAN9 expression showed a poor prognosis (Log-rank P<0.05). In addition, we detected that TSPAN9 was significantly decreased in HCC cells (P<0.01). As compared to the control cells, overexpression of TSPAN9 in HCC cells significantly reduced cell proliferation, slowed the wound healing rate, and inhibited the invasive and migration ability (all P<0.05). TCGA database analysis revealed a relationship between the expression of TSPAN9 and epithelium-mesenchymal transformation (EMT) factors.
Conclusions: Downregulated expression of TSPAN9 indicates a poor prognosis of HCC patients. TSPAN9 can inhibit the proliferation and metastasis of HCC cells.
Keywords: Hepatocellular carcinoma (HCC); metastasis; prognosis; proliferation; tetraspanin 9 (TSPAN9).
Copyright © 2025 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-2025-174/coif). The authors have no conflicts of interest to declare.
Figures


Similar articles
-
Nuclear factor IA-mediated transcriptional regulation of crystallin αB inhibits hepatocellular carcinoma progression.Mol Clin Oncol. 2025 Jun 20;23(2):72. doi: 10.3892/mco.2025.2867. eCollection 2025 Aug. Mol Clin Oncol. 2025. PMID: 40599718 Free PMC article.
-
[CENPI promotes the migration of liver cancer cells and the epithelial-mesenchymal transition process by activating the RAS/MEK/ERK signaling axis].Zhonghua Gan Zang Bing Za Zhi. 2025 Jul 20;33(7):674-682. doi: 10.3760/cma.j.cn501113-20231110-00189. Zhonghua Gan Zang Bing Za Zhi. 2025. PMID: 40784804 Chinese.
-
Mechanism of METTL14 regulates HBV-HCC malignant progression by mediating m6A modification of FOXP3 and thus transcriptional activation of ALDOB.J Mol Histol. 2025 Aug 8;56(4):259. doi: 10.1007/s10735-025-10551-y. J Mol Histol. 2025. PMID: 40778958
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
References
-
- Zhou Z, Li Q, Yu J, et al. Analysis of the epidemiological characteristics and disease burden of malignant tumors in Guangxi, 2019. Chinese Journal of Oncology Prevention and Treatment 2024;16:66-75.
LinkOut - more resources
Full Text Sources
Research Materials
